Fig. 3.
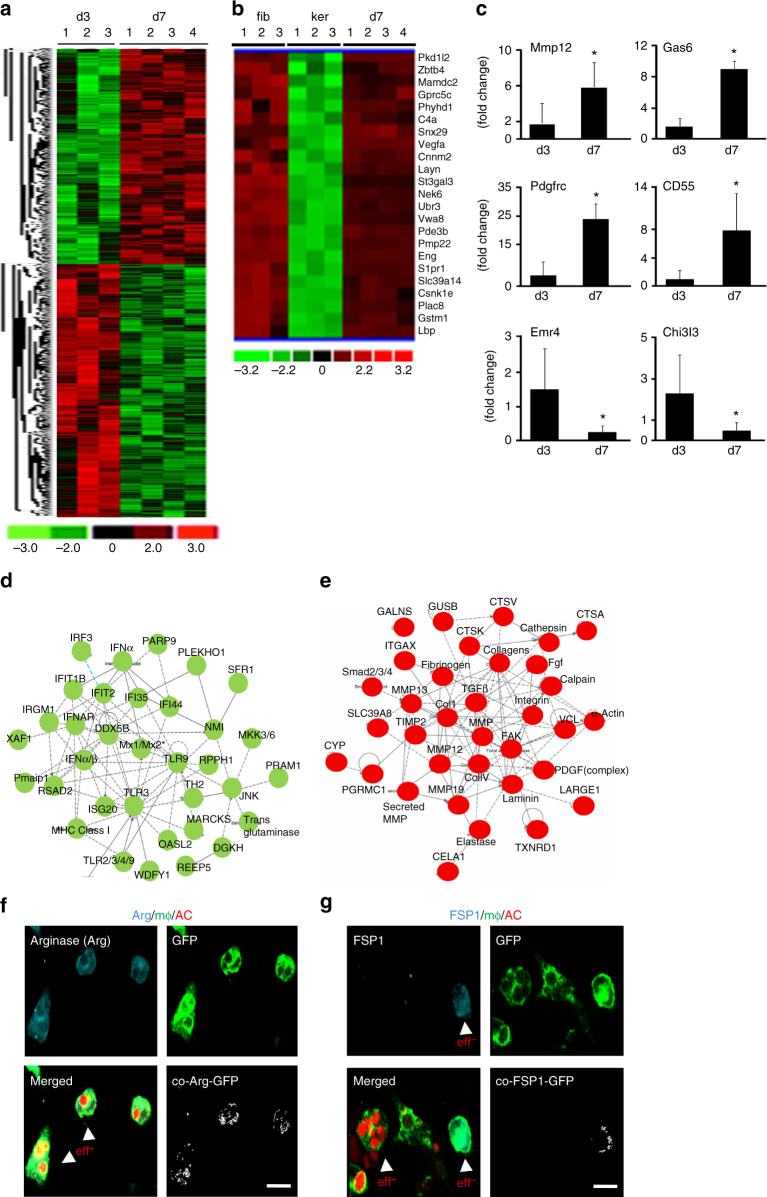
Transcriptome profiling of macrophage-derived fibroblast-like cells. a Heat map illustrating cluster of transcripts from CD11b+ F4/80+ wound macrophages at d3 and d7 post-wounding (PW). A total of 2757 genes were differentially expressed (p < 0.05, FDR). b Hierarchical cluster analysis of primary murine dermal fibroblast (GEO accession numbers: GSM 106139, GSM 106141, GSM 106149) and murine primary keratinocytes (GEO accession numbers: GSM 844759, GSM 844760, GSM 844761) transcriptomes obtained from Gene Expression Omnibus (GEO) database-enabled identification of a clear subset of fibroblast-specific genes differentially expressed in d7 CD11b+ F4/80+ wound macrophages. c qRT-PCR analysis of Mmp12, Gas6, PdgfrC, CD55, Emr4, and Chi3l3 mRNA expression identified differentially expressed in the transcriptome analysis, n = 5, Student’s t test p < 0.05. d, e Ingenuity® pathway analysis of the differentially expressed transcriptome. Pathways were scored using Fischer exact test (p < 0.05). d Downregulation of innate immune and inflammatory response genes in d7 wound macrophages compared to d3 wound macrophages. e Upregulation of fibroblast-specific genes in d7 wound macrophages. Pathways were scored using Fischer exact test (p < 0.05). f, g Efferocytosis with d3 wound macrophages (GFP+, green) from LysMCreRosamT/mG isolated and co-cultured with apoptotic cells (red). f Post-efferocytosis of apoptotic cell thymocytes (AC, red), wound macrophages (GFP, green) expressed arginase (cyan), n = 5, whereas g wound macrophages that did not participate in efferocytosis expressed FSP1 (cyan), n = 5, scale bar = 10 µm