Abstract
The Wnt signaling pathway plays a crucial role in neurodevelopment and in regulating the function and structure of the adult nervous system. Schizophrenia (SCZ) and bipolar disorder (BD) are severe mental disorders with evidence of subtle neurodevelopmental, structural and functional neuronal abnormalities. We aimed to elucidate the role of aberrant regulation of the Wnt system in these disorders by evaluating plasma levels of secreted Wnt modulators in patients (SCZ = 551 and BD = 246) and healthy controls (HCs = 639) using enzyme immune-assay. We also investigated the expression of 141 Wnt-related genes in whole blood in a subsample (SCZ = 338, BD = 241, and HCs = 263) using microarray analysis. Both SCZ and BD had dysregulated mRNA expression of Wnt-related genes favoring attenuated canonical (beta-catenin-dependent) signaling, and there were also indices of enhanced non-canonical Wnt signaling. In particular, FZD7, which may activate all Wnt pathways, but favors non-canonical signaling, and NFATc3, a downstream transcription factor and readout of the non-canonical Wnt/Ca2+ pathway, were significantly increased in SCZ and BD (p < 3 × 10−4). Furthermore, patients had lower plasma levels of soluble dickkopf 1 and sclerostin (p < 0.01) compared with HC. Our findings suggest that SCZ and BD are characterized by abnormal Wnt gene expression and plasma protein levels, and we propose that drugs targeting the Wnt pathway may have a role in the treatment of severe mental disorders.
Introduction
Schizophrenia (SCZ) and bipolar disorder (BD) are severe mental illnesses that are leading causes of worldwide disability1,2 and are associated with shortened lifespan3–5. Early brain morphological findings (i.e., aberrant neural lamination and orientation in the hippocampi), behavioral and cognitive alterations, and nonspecific motor development abnormalities in SCZ were among the first observations that underpinned the neurodevelopmental hypothesis of SCZ6. In BD, subtle brain morphological changes, behavioral alterations prior to onset of illness, and attenuations in neurogenesis may indicate disrupted neurodevelopment7. Further, in the adult brain, structural alterations, aberrant brain connectivity, and biochemical changes suggest patterns of subtle structural and functional brain abnormalities in both illnesses8,9. In addition, some studies have also indicated signs of neurodegenerative processes in SCZ10, with weaker evidence in BD11.
During neurodevelopment and in the adult brain the Wnt signaling pathway plays a crucial role in neural stem cell proliferation, differentiation and migration, neuroplasticity, and neurogenesis12,13. Secreted modulators such as the dickkopf (DKKs) and the secreted frizzled related proteins (sFRPs) regulate both the canonical (β-catenin-dependent) and non-canonical (β-catenin-independent) Wnt signaling, modulating the effects of various Wnt ligands14. Initial studies investigating the Wnt signaling pathway in severe mental disorders almost two decades ago focused on the hippocampus, where immunoreactivity of the Wnt ligand Wnt-1 was observed15. Aberrant findings in SCZ included reduced β-catenin16, increased Wnt-1 expression15, and reduced glycogen synthase kinase 3 beta (GSK-3β), a multifaceted kinase in Wnt signaling17. Later, large genome-wide association studies identified several polymorphisms in Wnt signaling in SCZ and BD suggesting a strong genetic component18. In particular, secreted Wnt antagonists, such as DKK419, sFRP1, and FZD320,21, have been associated with brain volumes22 and increased susceptibility to SCZ. In addition, these Wnt antagonists are located in the chromosomal region 8p, which is a genetically implicated region in neuropsychiatric disorders18. Furthermore, downstream modulators of β-catenin-dependent signaling have also been linked to increased susceptibility to both SCZ23,24 and BD, as well as suicidal behavior25 and body composition26. The Wnt signaling pathway is also linked to BD through investigations identifying that lithium impairs GSK-3β expression27. Although alterations in the Wnt signaling pathway have been evident for the past couple of decades a thorough understanding is still lacking. In particular, whereas most studies on the potential involvement of Wnt signaling in severe mental disorders have focused the canonical pathway, few studies has investigated the role of the non-canonical pathways that may be equally relevant in the pathogenesis of psychiatric disorders14. Furthermore, no studies have investigated serum levels of central secreted Wnt antagonists such as DKKs or frizzled related proteins.
In the present study we aimed to further elucidate the aberrations in the Wnt signaling pathway by conducting a pathway analysis on leukocyte mRNA in a large sample of SCZ and BD patients, and healthy controls (HCs) to identify differentially expressed genes as well as comparing circulating levels of secreted Wnt agonists and antagonists. We also aimed to explore associations between the Wnt signaling pathway and the use of medication.
Subjects and methods
Study design and ethics
The TOP Study at the NORMENT Centre, Oslo University Hospital, and collaborating Norwegian hospitals28 is approved by the Regional Committee for Medical Research Ethics and the Norwegian Data Inspectorate. The biobank is approved by the Norwegian Directorate of Health. All participants provided written informed consent after receiving a complete description of the study.
Participants
The main inclusion criteria were having a Diagnostic and Statistical Manual of Mental Disorders-IV (DSM-IV) diagnosis of SCZ spectrum disorders or bipolar spectrum disorders, intelligence quotient > 70, and age between 18 and 65 years (for details see ref. 28). Healthy volunteers without any history of severe psychiatric disorders (or in any of their first-degree relatives) or substance/alcohol abuse/dependency from the same catchment area were randomly selected from the National Population Registry (www.ssb.no). For the present analyses, patients and controls were not included if they had coexisting autoimmune or inflammatory disease, cancer, ongoing infections, used anti-inflammatory drugs, or had C-reactive protein levels above 20 mg/L. In total, 1625 participants included in the study had available plasma for protein assessment. Of these, 1436 participants (551 SCZ, 246 BD, and 639 HCs) met criteria for inclusion. A subsample of patients and controls (338 SCZ, 241 BD, and 263 HC participants) had available blood samples and passed quality control for microarray analyses.
Clinical assessments
Diagnosis was obtained using the Structured Clinical Interview for DSM-IV Axis I Disorders29. Clinical symptoms were evaluated using the Young Mania Rating Scale30, Inventory of Depressive Symptoms31, and Positive and Negative Syndrome Scale32, while functioning was measured using the Global Assessment of Functioning split version function and symptom scale33. The clinical assessment team consisted of psychologists and physicians who were all trained until satisfactory inter-rater reliability was obtained34. The use of psychotropic and other medication was also recorded at the time of inclusion. This was based on patient interview and medical records. On the day of blood sampling, the medication list was reconfirmed by asking patients about medication adherence (for details see ref. 35), and we also measured serum levels of medication from the same blood sample that was used for gene expression and protein level analyses.
Plasma protein assessment
We have previously described the blood sampling method36. We selected stabile plasma ligands that circulate at a detectable level: DKK1; DKK3; sclerostin (SOST), R-spondin-3 (RSPO3) and sFRP3. We measured their plasma levels in duplicate using commercially available antibodies (R&D Systems, Abingdon, UK) in a 384 format using a combination of a SELMA (Jena, Germany) pipetting robot and a BioTek (Winooski, VT, USA) dispenser/washer. For details see supplementary text 1.
RNA extraction
Blood samples were collected using Tempus Blood RNA Tubes. Total RNA was extracted with ABI PRISM 6100 Nucleic Acid PrepStation and TEMPUS 12-port RNA Isolation Kit according to the manufacturer’s protocol. High-Capacity cDNA Reverse Transcription Kit was used for reverse transcription of 1 µg RNA.
Global transcriptomics analyses
We selected 141 Wnt pathway-related genes using the Kyoto Encyclopedia of Genes and Genomes database (http://www.genome.jp/kegg/pathway.html). For each sample, 200 ng of total RNA was biotin-labeled and amplified using the Illumina TotalPrep-96 RNA Amplification Kit (Thermo Fisher, Waltham, MA, USA). Global analysis of gene expression was performed with Illumina HumanHT-12 v4 Bead Chip (Illumina, San Diego, CA, USA) consisting of more than 47 000 probes (i.e., transcripts). For this purpose, 842 samples (263 HCs, 338 SCZ, and 241 BD) passed labeling and scanning. Raw microarray scan files were exported using the Illumina GenomeStudio software and loaded into R for downstream analysis using specific packages provided by BioConductor37. Lumi was used to detect outliers38. R package (version 3.24.4.) was used to correct for technical batch effects, like RNA extraction batch, RNA extraction method, DNase treatment batch, cRNA labeling batch, and chip hybridization. Further quality control, quantile-normalization, and log2-transformation were done using Limma39.
Statistical analysis
Statistical analyses were performed using the SPSS software package for Windows, version 24.0 for plasma analyses. Data normality was assessed using the Kolmogorov–Smirnov and Shapiro–Wilk tests. We investigated differences in demographic data between groups using the chi-square test for categorical variables, the Kruskal–Wallis test for continuous variables, and the Mann–Whitney U-test for post hoc analyses. We used T-tests for normally distributed variables, and non-parametric tests (Mann–Whitney U-test) for skewed distributions to investigate differences between groups. Correlations were examined by using Spearman’s rank correlation. We controlled for age and sex in linear regression models.
To find associations between expression and the diagnostic group, a linear model was fitted in the R software environment using age, sex, and the expression level of Bmal1 as covariates. Bmal1 was included in the analyses to adjust for differences in time of blood sampling and circadian rhythm between patients and controls.
To explore possible associations between medication and the Wnt pathway we first calculated the defined daily dose of psychotropic medications (antipsychotics, mood stabilizers, and antidepressants) according to the guidelines from the World Health Organization Collaborating Center for Drug Statistics Methodology (https://www.whocc.no/atcdd). The defined daily dose is the assumed average maintenance dose per day for a drug used for its main indication in adults. We used serum concentration levels for lithium instead of defined daily dose. We selected significantly regulated Wnt antagonists in plasma (i.e., DKK1 and SOST) or major upstream or downstream differentially expressed genes (i.e., FZD7 or NFATc3). Associations were explored using analysis of covariance where we controlled for age, sex, and other medication groups, and we also investigated whether medication groups would influence these Wnt members (group effects; Supplementary table 2).
We corrected for multiple testing according to the Bonferroni method and alpha was set at p < 3 × 10−4 for our mRNA analyses differentially expressed genes (investigating 141 genes) and p < 0.01 for plasma ligand analyses (adjusting for 5 tests).
Tissue expression
In addition, we evaluated the tissue expression of FZD7 using the publically available Genotype-Tissue Expression (GTEx) dataset. The GTEx Project was supported by the Common Fund of the Office of the Director of the National Institutes of Health, and by NCI, NHGRI, NHLBI, NIDA, NIMH, and NINDS. The data used for the analyses described in this manuscript were obtained from [https://gtexportal.org/home/gene/FZD7] the GTEx Portal on 06/25/2017 and/or dbGaP accession number phs000424.vN.pN on 06/25/2017. NFATC3 protein expression in the brain was evaluated using The Human Protein Atlas (www.proteinatlas.org)40.
Results
Demographics and clinical characteristics
The socio-demographic and clinical characteristics of the participants are presented in Table 1. There were significant differences in ethnicity (more Caucasians in the HC group), sex (more males in the SCZ and HC group compared to BD), and age (BD and HC older than SCZ) between the patient groups and HCs. These differences were similar in the plasma (n = 1436) and the leukocyte (n = 842) cohorts. Patients with SCZ had higher symptom load and lower functioning than BD.
Table 1.
Demographic and clinical characteristics of participants
| Parameters | Plasma (Wnt ligand) cohort | Leukocyte (mRNA) cohort | ||||||
|---|---|---|---|---|---|---|---|---|
| SCZ (N = 551) | BD (N = 246) | HC (N = 639) | Post hoc analysis | SCZ (N = 338) | BD (N = 241) | HC (N = 263) | Post hoc analysis | |
| Male sex, N (%) | 334 (60.6) | 97 (39.4) | 364 (57.0) | SCZ > HC > BD | 206 (60.9) | 94 (39.0) | 144 (54.8) | SCZ > HC > BD |
| Ethnicity (Cauc. %) | 444 (80.6) | 213 (86.6) | 629 (98.4) | HC > BD > SCZ | 165 (90.7) | 141 (94.6) | 208 (100) | HC > BD > SCZ |
| Medication | ||||||||
| Antipsychotics | 510 (84.6) | 167 (66.0) | — | SCZ > BD | 172 (94.5) | 115 (77.2) | — | SCZ > BD |
| Lithium | 12 (2.0) | 51 (20.2) | — | BD > SCZ | 3 (1.2) | 32 (21.6) | — | BD > SCZ |
| Antidepressants | 179 (31.5) | 95 (38.8) | — | BD > SCZ | 51 (28.0) | 58 (38.9) | — | BD > SCZ |
| Mood stabilizers | 56 (9.3) | 87 (34.4) | — | BD > SCZ | 28 (15.4) | 66 (44.3) | — | BD > SCZ |
| Age (years) | 27 (13) | 29 (18) | 31 (13) | BD, HC > SCZ | 25 (11) | 36 (20) | 36.0 (11) | BD, HC > SCZ |
| DOI (years) | 4 (8) | 4 (10) | — | BD > SCZ | 5 (9) | 5 (13) | — | NS |
| PANSS total score | 62 (22) | 44 (13) | — | SCZ > BD | 64 (24) | 45 (14) | — | SCZ > BD |
| YMRS total score | 3 (9) | 2 (5) | — | SCZ > BD | 5 (8) | 2 (7) | — | SCZ > BD |
| IDS total score | 17 (19) | 17 (16) | — | NS | 19 (17) | 15 (17) | — | NS |
| GAF-S | 40 (15) | 57 (16) | — | BD > SCZ | 39 (10) | 54 (17) | — | BD > SCZ |
| GAF-F | 42 (14) | 51 (19) | — | BD > SCZ | 40 (11) | 50 (17) | — | BD > SCZ |
SCZ schizophrenia, BD bipolar disorder, HC healthy controls, Cauc. Caucasians, NS nonsignificant, DOI duration of illness, PANSS Positive and Negative Syndrome Scale, YMRS Young Mania Rating Scale, IDS Inventory of Depressive Symptoms, GAF-S Global Assessment of Functioning-Symptom Scale, GAF-F Global Assessment of Functioning-Function Scale
Categorical data are given as percent in brackets, while continuous data are given as median with interquartile range. Post hoc analysis is performed using Pearson chi-square for categorical data, and Mann–Whitney U-tests for continuous data
Plasma levels of Wnt modulators
The plasma levels of Wnt ligands in absolute values and group comparisons are summarized in Table 2a. Compared with HCs, patients had significantly lower levels of the Wnt antagonist DKK1 (p < 0.01) and SOST (p < 0.01 for SCZ and BD), also significant in adjusted analyses (i.e., age and sex). In the SCZ group, the levels of the Wnt antagonist sFRP3 were nominally decreased compared to HC (p = 0.04), with no significant differences in plasma ligand levels between SCZ and BD. As for the Wnt agonist RSPO3, we found nominally significant lower levels in SCZ, and no significant differences in BD compared to HC.
Table 2a.
Differences between patients and controls in Wnt pathway-related ligands
| Plasma ligands | M (IQR) | SCZ vs. HC | BD vs. HC | SCZ vs. BD | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| SCZ | BD | HC | n | z | t | n | z | t | n | z | t | |
| DKK1 | 1.63 (1.76) | 1.53 (1.63) | 1.99 (2.08) | 1245 | −4.08*** | −4.53*** | 895 | −4.37*** | — | 856 | 0.54 | 0.81 |
| DKK3 | 27.10 (9.01) | 27.80 (8.80) | 26.95 (9.02) | 1240 | −0.64 | −0.87 | 893 | 1.31 | 1.07 | 853 | −1.69 | −1.03 |
| sFRP3 | 3.45 (2.34) | 3.56 (2.64) | 3.88 (2.53) | 1245 | −2.64** | −2.10* | 895 | −1.26 | −0.71 | 856 | −0.69 | −1.23 |
| SOST | 134.11 (75.39) | 130.97 (68.24) | 148.93 (75.39) | 1245 | −4.13*** | −3.86*** | 895 | −3.11** | −2.67** | 856 | −0.13 | |
| RSPO3 | 70.9 (49.4) | 76.9 (47.6) | 76.4 (50.6) | 1245 | −2.51* | −2.03* | 895 | −0.80 | −0.87 | 856 | −1.07 | −0.72 |
Results are given as z from Mann–Whitney U-test and t from ANCOVA (linear regression analysis) controlling for age and gender. Results are significant after Bonferroni correction if p < 0.01, and nominally significant if 0.01 < p < 0.05
M median, IQR interquartile range, SCZ schizophrenia, BD bipolar disorder, HC healthy control, DKK1 dickkopf 1, DKK3 dickkopf 3, sFRP3 secreted frizzled related protein 3, SOST sclerostin, RSPO3 R-spondin-3
*p < 0.05; **p < 0.01; ***p < 0.0003
Differently expressed genes in SCZ and BD
The expression of Wnt pathway genes in whole blood are summarized in Figs. 1 and 2. Genes with significant differential expression (p < 3 × 10−4) are described in Table 2b, and nominal significant findings (3 × 10−4 < p < 0.05) in Supplementary table 1. Effect size estimates ranged between |0.03| and |0.15| in significant findings (p < 3 × 10−4), and between |0.02| and |0.13| in trend level findings (3 × 10−4 < p < 0.05).
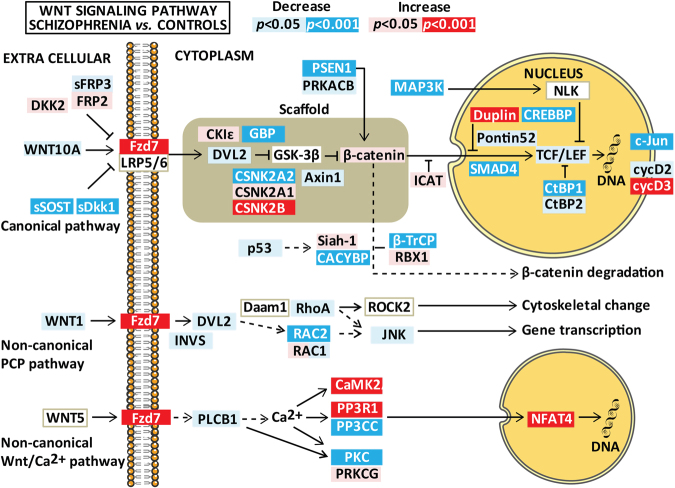
Fig. 1. Differences in Wnt pathway gene expression between patients with schizophrenia spectrum disorder and healthy controls after controlling for age, gender, and Bmal1.
Results are given as p-values where significant results (p-values adjusted for multiple testing) are indicated in red/dark blue (for increased/decreased mRNA expression) and nominally significant results (0.001 < p < 0.05) are shown as pink/light blue (for increased/decreased mRNA expression). The figure summarizes relevant genes for the three Wnt pathways (i.e., canonical pathway, non-canonical planar cell polarity pathway, and the non-canonical Wnt/Ca2+ pathway). The figure is based on the Wnt signaling pathway in the KEGG database
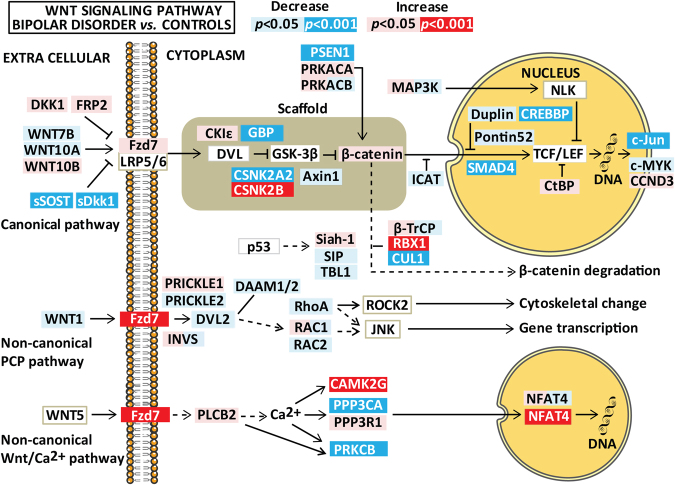
Fig. 2. Differences in Wnt pathway gene expression between patients with bipolar spectrum disorder and healthy controls after controlling for age, gender, and Bmal1.
Results are given as p-values where significant results (p-values adjusted for multiple testing) are indicated in red/dark blue (for increased/decreased mRNA expression) and nominally significant results (0.001 < p < 0.05) are shown as pink/light blue (for increased/decreased mRNA expression). The figure summarizes relevant genes for the three Wnt pathways (i.e., canonical pathway, non-canonical planar cell polarity pathway, and the non-canonical Wnt/Ca2+ pathway). The figure is based on the Wnt signaling pathway in the KEGG database
Table 2b.
Significant differences between patients and controls in Wnt signaling pathway gene mRNA expression after controlling for age and gender in linear models, and correction for multiple testing
| Gene | Protein names | Specificity | SCZ vs. HC | BD vs. HC | SCZ vs. BD | |
|---|---|---|---|---|---|---|
| B | B | B | ||||
| Wnt canonical pathway | PSEN1 | Presenilin-1 | ++ | −0.03**** | −0.02*** | 0.01 |
| FZD7 | Frizzled-7 | ++ | 0.07**** | 0.07*** | 0.00 | |
| β-catenin destruction complex | ||||||
| FRAT2 | GSK-3-binding protein FRAT2 | +++ | −0.13**** | −0.11*** | 0.02 | |
| CSNK2A2 | Casein kinase II subunit alpha | ++ | −0.05*** | −0.05**** | −0.01 | |
| CSNK2B | Casein kinase II subunit beta | ++ | 0.07**** | 0.06**** | −0.02 | |
| β-catenin degradation complex | ||||||
| CACYBP | Calcyclin-binding protein isoform 2 | +++ | −0.09**** | −0.06** | 0.03 | |
| FBXW11 | F-box/WD repeat-containing protein 11 | ++ | −0.04**** | −0.02* | 0.02 | |
| CUL1 | Cullin-1 | ++ | −0.02 | −0.06**** | 0.05** | |
| RBX1 | E3 ubiquitin-protein ligase RBX1 | ++ | 0.06* | 0.13**** | −0.07* | |
| β-catenin nuclear regulation | ||||||
| CHD8 | Chromodomain-helicase-DNA-binding protein 8 | +++ | 0.13**** | 0.09*** | −0.04 | |
| CREBBP | CREB-binding protein | + | −0.10**** | −0.09**** | 0.01 | |
| SMAD4 | Mothers against decapentaplegic homolog 4 | + | −0.09**** | −0.06**** | −0.03* | |
| CTBP1 | C-terminal binding protein 1 | ++ | −0.03**** | −0.01 | −0.02* | |
| MAP3K7 | Mitogen-activated protein kinase kinase kinase 7 | + | −0.03**** | −0.03** | 0.01 | |
| Transcription targets | ||||||
| JUN | Transcription factor AP-1 | + | −0.10**** | −0.10**** | 0.00 | |
| CCND3 | G1/S-specific cyclin-D3 | + | 0.11**** | 0.07** | 0.04 | |
| Wnt non-canonical PCP pathway | RAC2 | Ras-related C3 botulinum toxin substrate 2 | + | −0.07**** | −0.05** | 0.03 |
| Wnt non-canonical Ca++ pathway | CAMK2A | Calcium/calmodulin-dependent protein kinase type II subunit alpha | + | 0.06**** | 0.02 | 0.04** |
| CAMK2G | Calcium/calmodulin-dependent protein kinase type II subunit gamma | + | 0.01 | 0.07**** | −.05**** | |
| PPP3R1 | Calcineurin subunit B type 1 | + | 0.15**** | 0.10** | 0.05 | |
| PPP3CA | Serine/threonine-protein phosphatase 2B catalytic subunit alpha isoform | + | −0.02 | −0.07**** | 0.04** | |
| PPP3CC | Serine/threonine-protein phosphatase 2B catalytic subunit gamma isoform | + | −0.06**** | −0.03 | −0.03 | |
| PRKCB | Protein kinase C beta type | + | −0.11**** | −0.08**** | −0.03 | |
| NFATC3 | Nuclear factor of activated T cells, cytoplasmic 3 | + | 0.04**** | 0.04**** | 0.00 | |
Gene names are listed according to the HUGO Gene Nomenclature Committee and preferred protein names are given in brackets.
Results are given as effect size estimates from the linear regression analysis after correction for age, sex and BMAL1 expression. Results are significant after Bonferroni correction if p < 3 × 10−4, and nominally significant if 3 × 10−4 < p < 0.05.
SCZ schizophrenia, BD bipolar disorder, HC healthy control, B unstandardized regression coefficient
*p < 0.05; **p < 0.01; ***p < 0.001; ****p < 3×10−4
In general, gene expression in patients compared to HCs was quite similar for SCZ and BD with more modest changes in BD. For example, the Wnt receptor FZD7 expression was significantly increased in SCZ compared to HC (p < 3 × 10−4) with similar regression coefficients, but trend level changes in BD (p < 0.001). No other frizzled receptors or Wnt ligands were significantly regulated in our patient population.
Differently expressed genes involved in canonical Wnt signaling
The gamma secretase complex protein PSEN1 was decreased in SCZ compared to HC with a similar trend level decrease in BD. Other differentially expressed genes in the canonical pathway included several members of the β-catenin destruction complex (FRAT2 decreased in SCZ, CSNK2A2 decreased in BD, and CSNK2B increased in both), degradation complex (CACYBP and FBXW11 decreased in SCZ, and CUL decreased and RBX1 increased in BD), and nuclear regulation of β-catenin (CHD8 increased in SCZ, CTBP1 and MAP3K7 decreased in SCZ, and CREBBP and SMAD4 decreased in both). In addition, the transcriptional target JUN was decreased in SCZ and BD while CCND3 was increased in SCZ. The only significant differentially expressed genes between SCZ and BD were increased ICAT and decreased CAMK2G mRNA expression in SCZ (Table 2b, Supplementary Figure 1).
Differently expressed genes involved in non-canonical Wnt signaling in whole blood
The only gene involved in the non-canonical planar cell polarity pathway that was differently expressed was decreased RAC2 in SCZ compared to HC (Table 2b). For the non-canonical Wnt/Ca2+ pathway, a complex expression pattern was observed for genes involved in calcium-dependent cell signaling. First, one of the major isoforms of CaM kinase, CAMK2A, and PPP3R1, belonging to the regulatory subunit of calcineurin (CaN) were increased in SCZ, and CAMK2G in BD. Second, isozymes belonging to the catalytic subunit of CaN and protein kinase C (PKC) were downregulated (PPP3CA decreased in BD, PPP3CC decreased in SCZ, and PPP3CB and PRKCB decreased in both). Finally, NFAT3C was increased in both SCZ and BD.
Evaluation of tissue expression of FZD7 and NFATC3 from public databases
Our findings so far suggest that upregulation of the Wnt receptor FZD7 and NFAT3C, an important transcriptional factor of the non-canonical Wnt signaling pathway, may be major characteristics of SCZ and BD. To further elucidate these issues, we investigated tissue expression of these proteins in human brain based on available databases. Evaluation of tissue expression of FZD7 using the GTEx portal revealed the highest expression of FZD7 in the cerebellum compared to other tissues (Supplementary Figure 2). Protein expression data for NFATC3 in the brain are available from the Human Protein Atlas and show medium to high expression in all cell types in the hippocampus, cerebral cortex, caudate, and cerebellum (Supplementary Figure 3).
Role of medication
We found a positive association between defined daily dose of antipsychotics and plasma levels of DKK1 (β = 0.13, p < 0.01; Supplementary table 2). However, patients using antipsychotics did not have higher levels of DKK1 compared to patients not using antipsychotics. Patients using antidepressants (n = 249) had significantly lower levels of SOST (t = −2.91, p < 0.01) and increased FZD7 expression (t = 2.21, p < 0.05).
Discussion
We show for the first time in a large sample that the mRNA expression of FZD7 and NFATC3, two highly relevant Wnt signaling pathway genes, is significantly increased in patients with severe mental disorders compared to HCs. In addition, our pathway analyses indicate attenuated Wnt canonical pathway in patients vs. controls. Our study is also the first to investigate serum levels of secreted Wnt modulators in SCZ and BD, demonstrating significantly aberrant levels of DKK1 and SOST.
Interpretation of differentially expressed genes from signaling pathways is challenging since most intracellular components are not exclusive, but shared by multiple pathways and possibly more dependent on phosphorylation status than transcription levels. They may however aid in identifying dysregulated elements of a pathway that can be evaluated in detail in following studies. A major finding of our study was increased FZD7 mRNA levels in SCZ, and to a lesser degree in BD compared to controls. Among the frizzled receptors, FZD7 is unique as it can activate all Wnt pathways with non-canonical signaling being predominant41 and also oligomerize with all other FZD receptors and further modulate Wnt signaling42. Typical non-canonical Wnt ligands such as WNT5A and WNT11 bind FZD7 and activate the planar cell polarity43,44 or the Ca2+ pathway43,45, shown to be involved in SCZ46. Although data on FZD7 signaling in neural tissues is sparse, FZD7 mRNA is detected in multiple regions in the brain with heavily enriched expression in the cerebellum, which is affected in SCZ47. If the dysregulated FZD7 signaling is also present in the neural tissues of patients with psychotic disorders, the promiscuity of FZD7 with regard to interactions with other receptors and multiple Wnt pathways make it an appealing candidate for further studies.
Our second major finding regarding gene expression was increased NFATC3 mRNA expression in patients. NFATC3 codes the NFAT4 protein, and data from the Human Protein Atlas shows that NFAT4 protein is strongly expressed in brain tissue. Importantly, a recent genome-wide association study identified NFAT4 as a susceptibility gene in SCZ48. In addition to its role in inflammation and activation of immune cells, which is highly relevant in SCZ and BD, NFAT activation may participate in neuronal apoptosis in the CNS49. Specifically, NFAT4 activation has been implicated in neuronal loss in the hippocampus after brain injury50, is induced in astroglia51 and may promote inflammatory responses52. Wu et al.53, using an approach similar to ours, recently reported a downregulation of NFATC3 in a substantially smaller SCZ population. This discrepancy could be related to cellular composition as Wu et al. studied peripheral blood mononuclear cells while our samples contain all leukocyte populations, including a large proportion of neutrophils (~70%), and NFAT is an important inducer of inflammatory responses in neutrophils54. Non-canonical Wnt ligands can trigger the Wnt/Ca2+ pathway leading to activation of calcium-sensitive enzymes, including CaN, Ca2+/calmodulin-regulated kinase II, and PKC, which may activate NFAT signaling55. We and others have demonstrated that non-canonical WNT5A stimulation enhanced NFAT activity56,57. Based on the complex expression pattern of CaN and PKC transcripts, but increased CAMK2 in SCZ and BD, it is tempting to hypothesize that CAMK2 could activate NFAT as has been demonstrated in lymphocytes58,59. Regardless of the mechanisms for upregulation, enhanced NFAT4 activity in the brain and/or immune cells could potentially induce neuro-inflammatory responses in SCZ and BD. Of relevance, both NFAT60 and CAMK261 may antagonize the canonical pathway.
In support of a previous study53 we found evidence of attenuated canonical Wnt signaling in SCZ and BD. This is suggested by decreased mRNA levels of members belonging to the β-catenin destruction complex (i.e., CSNK2A2 and FRAT2), essential for Wnt/β-catenin signaling62,63. Aberrant CK2 signaling in the cortex has been linked to altered neurotransmitter release in SCZ64. Decreased PSEN1 would favor enhanced canonical Wnt signaling, but may also have multiple functions outside of the γ-secretase complex. In addition, regulated levels of components involved in p-53-mediated degradation of β-catenin (e.g., the CACYBP, β-TrCP, CUL1, and RBX1) could reflect altered turnover of β-catenin, but are difficult to interpret as multiple non-Wnt substrates are processed by this complex. We identified several differentially expressed genes involved in nuclear transport and regulation of β-catenin, such as increased CHD8, a nuclear protein that inhibits β-catenin signaling65, and decreased MAP3K7. MAP3K7 is also known as TAK1, and studies in TAK(−/−) mice reveal that low levels may adversely affect cerebellar development and neuro-developmentally regulated behavior66. Decreased expression of transcriptional co-activators of canonical Wnt target genes such as CtBP1 (both co-repressor and co-activator), CREBBP, a protein linked to SCZ susceptibility67, and the transforming growth factor transcription factor SMAD4 are also compatible with attenuated canonical signaling. These finding are supported by decreased expression of the Wnt target gene JUN, which may further reinforce Wnt signaling68 and increased CCND3, which in contrast to cyclin D1/D2, is increased in response to inhibition of canonical Wnt signaling69,70.
Interpretation of signaling networks in cross-sectional studies is further complicated by that changes in expression may reflect compensatory feedback mechanisms in response to dysregulated signaling. Multiple families of secreted antagonists or modulators may regulate Wnt signaling71 and the decreased circulating levels of DKK1, an antagonist of the canonical Wnt pathway, observed in SCZ and BD could reflect a downregulation of DKK1in an attempt to enhance canonical Wnt signaling. In an analogous fashion, we have shown WNT5A stimulates the release of sFRP3, which antagonizes WNT5A-induced NFAT and ERK activity57,72. Alternatively, DKK1 may reflect decreased activity in the Wnt canonical pathway as DKK1 is directly regulated by the β-catenin/TCF complex under physiological circumstances73. Low serum levels of DKK1 have been associated with increased risk of somatic diseases, and to predict increased mortality in older patients74. SOST is also a potent inhibitor of the Wnt canonical pathway75, and low circulating levels are frequently seen in relation to enhanced calcification76, which is relevant as calcification of the choroid plexus has been associated with regional brain atrophies and neurodegenerative phenotype in SCZ and BD77. Serum levels of SOST have not been investigated in severe mental disorders, but are inversely correlated with vitamin D levels78, and low vitamin D associates with symptoms in severe mental disorders79.
We investigated whether our findings may be attributed to the use of medication. Due to the naturalistic nature of our study most patients were using a combination of psychotropic medication. We detected a significant dosage-dependent association between antipsychotics and DKK1, however we found no group effects of antipsychotics. SOST levels were lower in the group using antidepressants, which is compatible with an activation in canonical Wnt signaling80.
Our study has some limitations that should be considered. First, we did not measure phosphorylation status or protein levels of intracellular components of the Wnt pathways, thus our conclusions rely on gene expression data and plasma protein levels. Second, although a post hoc power evaluation revealed an observed power above 0.90 for most transcripts and circulating proteins, observed power was low (below 0.60), for several low-abundant transcripts. Third, many patients were using a combination of psychotropic medications, which made it impossible to investigate effects of medication in monotherapy. We did, however, control for co-medication in the statistical analyses. Lastly, the cross-sectional design of the study hinders us from making strong claims about causality.
In summary, we provide further evidence of altered Wnt signaling in SCZ and BD in a well-powered sample. In particular, we show that NFATC3 and FZD7 mRNA expression is increased in peripheral blood of patients, while they have lower serum levels of DKK1 and SOST compared to HCs. Our findings could suggest that drugs blocking the non-canonical Wnt Ca pathway (e.g., WNT5A antagonists) could have a role in the treatment of severe mental disorders and warrants further investigation.
Electronic supplementary material
Acknowledgements
We would like to thank the participants of the study. We are also grateful for the financial support provided by the KG Jebsen Stiftelsen, Research Council of Norway (223273 and 248778), and South East Norway Health Authority (2017-112) for this study. We also acknowledge the technical support and service from the Genomics Core Facility at the Department of Clinical Science, the University of Bergen.
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Electronic supplementary material
Supplementary Information accompanies this paper at 10.1038/s41398-018-0102-1.
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Ferrari AJ, et al. The prevalence and burden of bipolar disorder: findings from the Global Burden of Disease Study 2013. Bipolar Disord. 2016;18:440–450. doi: 10.1111/bdi.12423. [DOI] [PubMed] [Google Scholar]
- 2.Chong HY, et al. Global economic burden of schizophrenia: a systematic review. Neuropsychiatr. Dis. Treat. 2016;12:357–373. doi: 10.2147/NDT.S96649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Crump C, Sundquist K, Winkleby MA, Sundquist J. Comorbidities and mortality in bipolar disorder: a Swedish national cohort study. JAMA Psychiatry. 2013;70:931–939. doi: 10.1001/jamapsychiatry.2013.1394. [DOI] [PubMed] [Google Scholar]
- 4.Laursen TM, Nordentoft M, Mortensen PB. Excess early mortality in schizophrenia. Annu. Rev. Clin. Psychol. 2014;10:425–448. doi: 10.1146/annurev-clinpsy-032813-153657. [DOI] [PubMed] [Google Scholar]
- 5.Laursen TM, Munk-Olsen T, Vestergaard M. Life expectancy and cardiovascular mortality in persons with schizophrenia. Curr. Opin. Psychiatry. 2012;25:83–88. doi: 10.1097/YCO.0b013e32835035ca. [DOI] [PubMed] [Google Scholar]
- 6.Kozlovsky N, Belmaker RH, Agam G. GSK-3 and the neurodevelopmental hypothesis of schizophrenia. Eur. Neuropsychopharmacol. 2002;12:13–25. doi: 10.1016/S0924-977X(01)00131-6. [DOI] [PubMed] [Google Scholar]
- 7.O’Shea KS, McInnis MG. Neurodevelopmental origins of bipolar disorder: iPSC models. Mol. Cell. Neurosci. 2016;73:63–83. doi: 10.1016/j.mcn.2015.11.006. [DOI] [PubMed] [Google Scholar]
- 8.Hibar DP, et al. Subcortical volumetric abnormalities in bipolar disorder. Mol. Psychiatry. 2016;21:1710–1716. doi: 10.1038/mp.2015.227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.van Erp TG, et al. Subcortical brain volume abnormalities in 2028 individuals with schizophrenia and 2540 healthy controls via the ENIGMA consortium. Mol. Psychiatry. 2016;21:585. doi: 10.1038/mp.2015.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pino O, et al. Neurodevelopment or neurodegeneration: review of theories of schizophrenia. Actas Esp. Psiquiatr. 2014;42:185–195. [PubMed] [Google Scholar]
- 11.Savitz JB, Price JL, Drevets WC. Neuropathological and neuromorphometric abnormalities in bipolar disorder: view from the medial prefrontal cortical network. Neurosci. Biobehav. Rev. 2014;42:132–147. doi: 10.1016/j.neubiorev.2014.02.008. [DOI] [PubMed] [Google Scholar]
- 12.Meffre D, et al. Wnt and lithium: a common destiny in the therapy of nervous system pathologies? Cell. Mol. Life Sci. 2014;71:1123–1148. doi: 10.1007/s00018-013-1378-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Valvezan AJ, Klein PS. GSK-3 and Wnt signaling in neurogenesis and bipolar disorder. Front. Mol. Neurosci. 2012;5:1. doi: 10.3389/fnmol.2012.00001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Okerlund ND, Cheyette BN. Synaptic Wnt signaling-a contributor to major psychiatric disorders? J. Neurodev. Disord. 2011;3:162–174. doi: 10.1007/s11689-011-9083-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Miyaoka T, Seno H, Ishino H. Increased expression of Wnt-1 in schizophrenic brains. Schizophr. Res. 1999;38:1–6. doi: 10.1016/S0920-9964(98)00179-0. [DOI] [PubMed] [Google Scholar]
- 16.Cotter D, et al. Abnormalities of Wnt signalling in schizophrenia—evidence for neurodevelopmental abnormality. Neuroreport. 1998;9:1379–1383. doi: 10.1097/00001756-199805110-00024. [DOI] [PubMed] [Google Scholar]
- 17.Beasley C, et al. Glycogen synthase kinase-3beta immunoreactivity is reduced in the prefrontal cortex in schizophrenia. Neurosci. Lett. 2001;302:117–120. doi: 10.1016/S0304-3940(01)01688-3. [DOI] [PubMed] [Google Scholar]
- 18.Tabares-Seisdedos R, Rubenstein JL. Chromosome 8p as a potential hub for developmental neuropsychiatric disorders: implications for schizophrenia, autism and cancer. Mol. Psychiatry. 2009;14:563–589. doi: 10.1038/mp.2009.2. [DOI] [PubMed] [Google Scholar]
- 19.Proitsi P, et al. Positional pathway screen of wnt signaling genes in schizophrenia: association with DKK4. Biol. Psychiatry. 2008;63:13–16. doi: 10.1016/j.biopsych.2007.03.014. [DOI] [PubMed] [Google Scholar]
- 20.Katsu T, et al. The human frizzled-3 (FZD3) gene on chromosome 8p21, a receptor gene for Wnt ligands, is associated with the susceptibility to schizophrenia. Neurosci. Lett. 2003;353:53–56. doi: 10.1016/j.neulet.2003.09.017. [DOI] [PubMed] [Google Scholar]
- 21.Zhang Y, et al. Positive association of the human frizzled 3 (FZD3) gene haplotype with schizophrenia in Chinese Han population. Am. J. Med. Genet. B Neuropsychiatr. Genet. 2004;129b:16–19. doi: 10.1002/ajmg.b.30076. [DOI] [PubMed] [Google Scholar]
- 22.Zhang LH, et al. [Association of the schizophrenia susceptible gene DKK4 with brain volume in Chinese populations] Dongwuxue Yanjiu. 2011;32:62–65. doi: 10.3724/SP.J.1141.2011.01062. [DOI] [PubMed] [Google Scholar]
- 23.Nivard MG, et al. Further confirmation of the association between anxiety and CTNND2: replication in humans. Genes Brain Behav. 2014;13:195–201. doi: 10.1111/gbb.12095. [DOI] [PubMed] [Google Scholar]
- 24.Zhang J, Chen J, Xu Q, Shen Y. Does the presenilin 2 gene predispose to schizophrenia? Schizophr. Res. 2009;109:121–129. doi: 10.1016/j.schres.2009.01.013. [DOI] [PubMed] [Google Scholar]
- 25.Jimenez E, et al. Genetic variability at IMPA2, INPP1 and GSK3beta increases the risk of suicidal behavior in bipolar patients. Eur. Neuropsychopharmacol. 2013;23:1452–1462. doi: 10.1016/j.euroneuro.2013.01.007. [DOI] [PubMed] [Google Scholar]
- 26.Winham SJ, et al. Genome-wide association study of bipolar disorder accounting for effect of body mass index identifies a new risk allele in TCF7L2. Mol. Psychiatry. 2014;19:1010–1016. doi: 10.1038/mp.2013.159. [DOI] [PubMed] [Google Scholar]
- 27.Ferensztajn-Rochowiak E, Rybakowski JK. The effect of lithium on hematopoietic, mesenchymal and neural stem cells. Pharmacol. Rep. 2016;68:224–230. doi: 10.1016/j.pharep.2015.09.005. [DOI] [PubMed] [Google Scholar]
- 28.Dieset I, et al. Up-regulation of NOTCH4 gene expression in bipolar disorder. Am. J. Psychiatry. 2012;169:1292–1300. doi: 10.1176/appi.ajp.2012.11091431. [DOI] [PubMed] [Google Scholar]
- 29.First, M. B., Spitzer, R. L., Goibbon, M. & Williams, J. B. W. Structured Clinical Interview for DSM-IV Axis I Disorders: Patient Edition (SCID-P), Version 2 (New York State Psychiatric Institute, New York, 1995).
- 30.Young RC, Biggs JT, Ziegler VE, Meyer DA. A rating scale for mania: reliability, validity and sensitivity. Br. J. Psychiatry. 1978;133:429–435. doi: 10.1192/bjp.133.5.429. [DOI] [PubMed] [Google Scholar]
- 31.Rush AJ, Gullion CM, Basco MR, Jarrett RB, Trivedi MH. The Inventory of Depressive Symptomatology (IDS): psychometric properties. Psychol. Med. 1996;26:477–486. doi: 10.1017/S0033291700035558. [DOI] [PubMed] [Google Scholar]
- 32.Kay SR, Fiszbein A, Opler LA. The positive and negative syndrome scale (PANSS) for schizophrenia. Schizophr. Bull. 1987;13:261–276. doi: 10.1093/schbul/13.2.261. [DOI] [PubMed] [Google Scholar]
- 33.Pedersen G, Hagtvet KA, Karterud S. Generalizability studies of the Global Assessment of Functioning-Split version. Compr. Psychiatry. 2007;48:88–94. doi: 10.1016/j.comppsych.2006.03.008. [DOI] [PubMed] [Google Scholar]
- 34.Lagerberg TV, et al. Indications of a dose-response relationship between cannabis use and age at onset in bipolar disorder. Psychiatry Res. 2014;215:101–104. doi: 10.1016/j.psychres.2013.10.029. [DOI] [PubMed] [Google Scholar]
- 35.Jonsdottir H, et al. Medication adherence in outpatients with severe mental disorders: relation between self-reports and serum level. J. Clin. Psychopharmacol. 2010;30:169–175. doi: 10.1097/JCP.0b013e3181d2191e. [DOI] [PubMed] [Google Scholar]
- 36.Hope S, et al. Similar immune profile in bipolar disorder and schizophrenia: selective increase in soluble tumor necrosis factor receptor I and von Willebrand factor. Bipolar Disord. 2009;11:726–734. doi: 10.1111/j.1399-5618.2009.00757.x. [DOI] [PubMed] [Google Scholar]
- 37.Ritchie ME, Dunning MJ, Smith ML, Shi W, Lynch AG. BeadArray expression analysis using bioconductor. PLoS Comput. Biol. 2011;7:e1002276. doi: 10.1371/journal.pcbi.1002276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Du P, Kibbe WA, Lin SM. lumi: a pipeline for processing Illumina microarray. Bioinformatics. 2008;24:1547–1548. doi: 10.1093/bioinformatics/btn224. [DOI] [PubMed] [Google Scholar]
- 39.Ritchie ME, et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43:e47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Uhlen M, et al. Proteomics. Tissue-based map of the human proteome. Science. 2015;347:1260419. doi: 10.1126/science.1260419. [DOI] [PubMed] [Google Scholar]
- 41.Phesse T, Flanagan D, Vincan E. Frizzled7: a promising Achilles’ heel for targeting the Wnt receptor complex to treat cancer. Cancers (Basel) 2016;8:50. doi: 10.3390/cancers8050050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kaykas A, et al. Mutant Frizzled 4 associated with vitreoretinopathy traps wild-type Frizzled in the endoplasmic reticulum by oligomerization. Nat. Cell Biol. 2004;6:52–58. doi: 10.1038/ncb1081. [DOI] [PubMed] [Google Scholar]
- 43.De Calisto J, Araya C, Marchant L, Riaz CF, Mayor R. Essential role of non-canonical Wnt signalling in neural crest migration. Development. 2005;132:2587–2597. doi: 10.1242/dev.01857. [DOI] [PubMed] [Google Scholar]
- 44.Le Grand F, Jones AE, Seale V, Scime A, Rudnicki MA. Wnt7a activates the planar cell polarity pathway to drive the symmetric expansion of satellite stem cells. Cell Stem Cell. 2009;4:535–547. doi: 10.1016/j.stem.2009.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Winklbauer R, Medina A, Swain RK, Steinbeisser H. Frizzled-7 signalling controls tissue separation during Xenopus gastrulation. Nature. 2001;413:856–860. doi: 10.1038/35101621. [DOI] [PubMed] [Google Scholar]
- 46.Devor A, et al. Genetic evidence for role of integration of fast and slow neurotransmission in schizophrenia. Mol. Psychiatry. 2017;22:792–801. doi: 10.1038/mp.2017.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Moberget T., et al. Cerebellar volume and cerebellocerebral structural covariance in schizophrenia: a multisite mega-analysis of 983 patients and 1349 healthy controls. Mol Psychiatry (2017). 10.1038/mp.2017.106 [DOI] [PubMed]
- 48.Pouget JG, et al. Genome-wide association studies suggest limited immune gene enrichment in schizophrenia compared to 5 autoimmune diseases. Schizophr. Bull. 2016;42:1176–1184. doi: 10.1093/schbul/sbw059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gomez-Sintes R, Lucas JJ. NFAT/Fas signaling mediates the neuronal apoptosis and motor side effects of GSK-3 inhibition in a mouse model of lithium therapy. J. Clin. Invest. 2010;120:2432–2445. doi: 10.1172/JCI37873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yan HQ, Shin SS, Ma X, Li Y, Dixon CE. Differential effect of traumatic brain injury on the nuclear factor of activated T Cells C3 and C4 isoforms in the rat hippocampus. Brain Res. 2014;1548:63–72. doi: 10.1016/j.brainres.2013.12.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Serrano-Perez MC, et al. Response of transcription factor NFATc3 to excitotoxic and traumatic brain insults: identification of a subpopulation of reactive astrocytes. Glia. 2011;59:94–107. doi: 10.1002/glia.21079. [DOI] [PubMed] [Google Scholar]
- 52.Neria F, et al. NFATc3 promotes Ca(2+)-dependent MMP3 expression in astroglial cells. Glia. 2013;61:1052–1066. doi: 10.1002/glia.22494. [DOI] [PubMed] [Google Scholar]
- 53.Wu JQ, et al. Altered neural signaling and immune pathways in peripheral blood mononuclear cells of schizophrenia patients with cognitive impairment: a transcriptome analysis. Brain Behav. Immun. 2016;53:194–206. doi: 10.1016/j.bbi.2015.12.010. [DOI] [PubMed] [Google Scholar]
- 54.Fric J, et al. NFAT control of innate immunity. Blood. 2012;120:1380–1389. doi: 10.1182/blood-2012-02-404475. [DOI] [PubMed] [Google Scholar]
- 55.Kuhl M. The WNT/calcium pathway: biochemical mediators, tools and future requirements. Front. Biosci. 2004;9:967–974. doi: 10.2741/1307. [DOI] [PubMed] [Google Scholar]
- 56.Bradley EW, Drissi MH. WNT5A regulates chondrocyte differentiation through differential use of the CaN/NFAT and IKK/NF-kappaB pathways. Mol. Endocrinol. 2010;24:1581–1593. doi: 10.1210/me.2010-0037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Abraityte A, et al. Wnt5a is associated with right ventricular dysfunction and adverse outcome in dilated cardiomyopathy. Sci. Rep. 2017;7:3490. doi: 10.1038/s41598-017-03625-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Nghiem P, Ollick T, Gardner P, Schulman H. Interleukin-2 transcriptional block by multifunctional Ca2+/calmodulin kinase. Nature. 1994;371:347–350. doi: 10.1038/371347a0. [DOI] [PubMed] [Google Scholar]
- 59.Ishiguro K, et al. Ca2+/calmodulin-dependent protein kinase II is a modulator of CARMA1-mediated NF-kappaB activation. Mol. Cell. Biol. 2006;26:5497–5508. doi: 10.1128/MCB.02469-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Huang T, et al. Nuclear factor of activated T cells (NFAT) proteins repress canonical Wnt signaling via its interaction with Dishevelled (Dvl) protein and participate in regulating neural progenitor cell proliferation and differentiation. J. Biol. Chem. 2011;286:37399–37405. doi: 10.1074/jbc.M111.251165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ishitani T, et al. The TAK1-NLK mitogen-activated protein kinase cascade functions in the Wnt-5a/Ca(2+) pathway to antagonize Wnt/beta-catenin signaling. Mol. Cell. Biol. 2003;23:131–139. doi: 10.1128/MCB.23.1.131-139.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Saitoh T, et al. Molecular cloning and characterization of FRAT2, encoding a positive regulator of the WNT signaling pathway. Biochem. Biophys. Res. Commun. 2001;281:815–820. doi: 10.1006/bbrc.2001.4421. [DOI] [PubMed] [Google Scholar]
- 63.Gao Y, Wang HY. Casein kinase 2 Is activated and essential for Wnt/beta-catenin signaling. J. Biol. Chem. 2006;281:18394–18400. doi: 10.1074/jbc.M601112200. [DOI] [PubMed] [Google Scholar]
- 64.Castillo MA, Ghose S, Tamminga CA, Ulery-Reynolds PG. Deficits in syntaxin 1 phosphorylation in schizophrenia prefrontal cortex. Biol. Psychiatry. 2010;67:208–216. doi: 10.1016/j.biopsych.2009.07.029. [DOI] [PubMed] [Google Scholar]
- 65.Sakamoto I, et al. A novel beta-catenin-binding protein inhibits beta-catenin-dependent Tcf activation and axis formation. J. Biol. Chem. 2000;275:32871–32878. doi: 10.1074/jbc.M004089200. [DOI] [PubMed] [Google Scholar]
- 66.Kim YS, et al. Altered cerebellar development in nuclear receptor TAK1/ TR4 null mice is associated with deficits in GLAST(+) glia, alterations in social behavior, motor learning, startle reactivity, and microglia. Cerebellum. 2010;9:310–323. doi: 10.1007/s12311-010-0163-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Crisafulli C, et al. Possible influence of CREB1, CREBBP and CREM variants on diagnosis and treatment outcome in patients with schizophrenia. Neurosci. Lett. 2012;508:37–41. doi: 10.1016/j.neulet.2011.12.013. [DOI] [PubMed] [Google Scholar]
- 68.Gan XQ, et al. Nuclear Dvl, c-Jun, beta-catenin, and TCF form a complex leading to stabilization of beta-catenin-TCF interaction. J. Cell Biol. 2008;180:1087–1100. doi: 10.1083/jcb.200710050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Keats EC, Dominguez JM, 2nd, Grant MB, Khan ZA. Switch from canonical to noncanonical Wnt signaling mediates high glucose-induced adipogenesis. Stem Cells. 2014;32:1649–1660. doi: 10.1002/stem.1659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Tanaka S, Terada K, Nohno T. Canonical Wnt signaling is involved in switching from cell proliferation to myogenic differentiation of mouse myoblast cells. J. Mol. Signal. 2011;6:12. doi: 10.1186/1750-2187-6-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kawano Y, Kypta R. Secreted antagonists of the Wnt signalling pathway. J. Cell Sci. 2003;116:2627–2634. doi: 10.1242/jcs.00623. [DOI] [PubMed] [Google Scholar]
- 72.Abraityte A, et al. Wnt5a is elevated in heart failure and affects cardiac fibroblast function. J. Mol. Med. 2017;95:767–777. doi: 10.1007/s00109-017-1529-1. [DOI] [PubMed] [Google Scholar]
- 73.Niida A, et al. DKK1, a negative regulator of Wnt signaling, is a target of the beta-catenin/TCF pathway. Oncogene. 2004;23:8520–8526. doi: 10.1038/sj.onc.1207892. [DOI] [PubMed] [Google Scholar]
- 74.Dovjak P, Heinze G, Rainer A, Sipos W, Pietschmann P. Serum levels of Dickkopf-1 are a potential negative biomarker of survival in geriatric patients. Exp. Gerontol. 2017;96:104–109. doi: 10.1016/j.exger.2017.06.002. [DOI] [PubMed] [Google Scholar]
- 75.Hay E, Bouaziz W, Funck-Brentano T, Cohen-Solal M. Sclerostin and bone aging: a mini-review. Gerontology. 2016;62:618–623. doi: 10.1159/000446278. [DOI] [PubMed] [Google Scholar]
- 76.Evenepoel P, et al. Sclerostin serum levels and vascular calcification progression in prevalent renal transplant recipients. J. Clin. Endocrinol. Metab. 2015;100:4669–4676. doi: 10.1210/jc.2015-3056. [DOI] [PubMed] [Google Scholar]
- 77.Yarlagadda A, Kaushik S, Clayton AH. Blood brain barrier: the role of calcium homeostasis. Psychiatry. 2007;4:55–59. [PMC free article] [PubMed] [Google Scholar]
- 78.Ardawi MS, Al-Kadi HA, Rouzi AA, Qari MH. Determinants of serum sclerostin in healthy pre- and postmenopausal women. J. Bone Mineral. Res. 2011;26:2812–2822. doi: 10.1002/jbmr.479. [DOI] [PubMed] [Google Scholar]
- 79.Nerhus M, et al. Low vitamin D is associated with negative and depressive symptoms in psychotic disorders. Schizophr. Res. 2016;178:44–49. doi: 10.1016/j.schres.2016.08.024. [DOI] [PubMed] [Google Scholar]
- 80.Sani G, et al. The wnt pathway in mood disorders. Curr. Neuropharmacol. 2012;10:239–253. doi: 10.2174/157015912803217279. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.