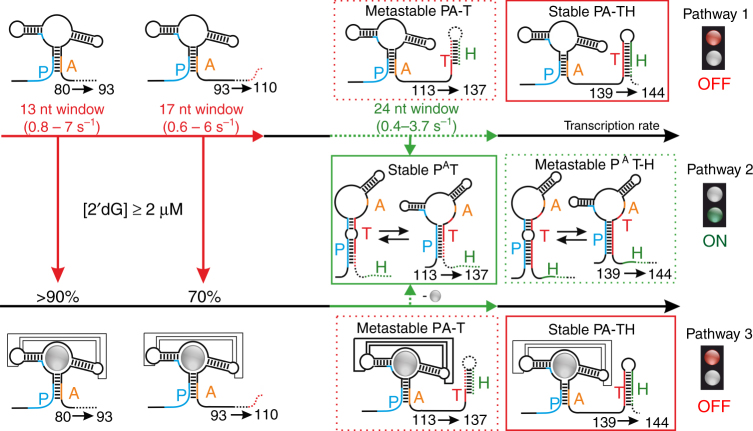
Fig. 1.
Non-equilibrium model for the co-transcriptional conformational switch of the 2′dG-sensing riboswitch. Regulatory function by transcriptional riboswitches is based on a ligand-dependent interplay between the four sequence segments P (5′-aptamer strand), A (aptamer-stabilizing strand), T (switching strand), and H (terminator strand)15. Previously derived transcription intervals for folding events involving a stabilization of the aptamer domain via the P–A interaction or switch in strand interaction (A–T interaction) are highlighted in red (ligand binding) and green (ON state folding), respectively. Directly after transcription of the aptamer domain, 2′dG can bind during the synthesis of nts 80–110 (30 nt window), with a continuous decrease in the ligand binding efficiency from >90% to 70% between nucleotides 93 and 110. Continuation of transcription beyond nt 113 transforms both the ligand-bound and ligand-free aptamer domains (PA–T) into metastable states (dashed red box). Folding to the regulatory ON state (PAT, solid green box) can only occur during synthesis of nucleotides 113–137. At the regulatory decision point, folded PAT–H states are metastable (dashed green box), while the PA–TH conformation (solid red box) represents the lowest free energy state in both absence and presence of ligand. Metastable states are highlighted by dashed boxes and lowest free energy states by solid boxes