Fig. 6.
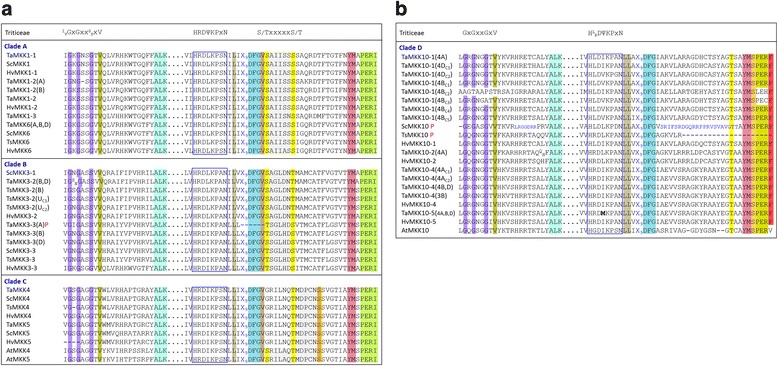
Alignment of important domains of Triticeae MKKs. (a) clades A, B and C, and (b) clade D MKKs. The amino acid sequences from multiple genomic copies marked in bold in Additional file 3 were aligned in Clustal Omega (www.ebi.ac.uk/Tools/msa/clustalo/) using default settings. The conserved domains are highlighted in different colours. The conserved P-loop anchor found in the ATP binding pocket of PKs (GxGxxG) and the C-loop (DΨK consensus; where Ψ refers to the aliphatic amino acids [79] L/I/V, but not M) are represented above the alignment. All genomic copies of a given MKK from wheat or triticale are represented together. Superscript/subscripts are used where there is sequence variation among copies. In some cases, due to variability among sequences, multiple copies of wheat and triticale are represented. Letters in parenthesis after names indicate the source genome. U indicates unidentified chromosome number. The subscripts (C1….C5) at the end of the MKK names indicate copy numbers. The bold letters represent deviation from preferred amino acids at that position. Smaller font size (blue font) was used for some sequences in clade D for alignment adjustment purposes. The letter P (red font) refers to accessions for which only a partial sequence is available
