SUMMARY
Cryptococcus neoformans (C. neoformans) is estimated to cause about 220,000 new cases every year in patients with AIDS, despite advances in antifungal treatments. C. neoformans possesses a remarkable ability to disseminate through an immunocompromised host, making treatment difficult. Here, we examine the mechanism of survival of C. neoformans under varying host conditions and find a role for ceramide synthase in C. neoformans virulence. This study also provides a detailed lipidomics resource for the fungal lipid research community in addition to discovering a potential target for antifungal therapy.
In Brief
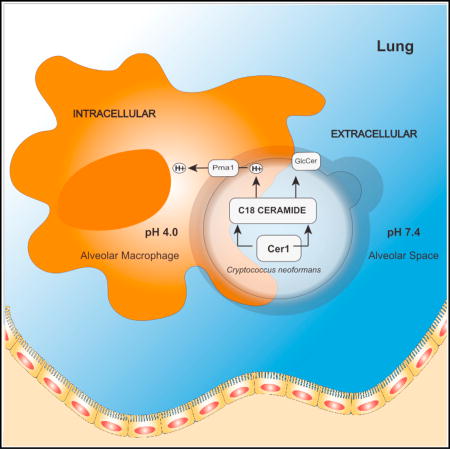
Cryptococcus neoformans (C. neoformans) is a fungal pathogen that causes about 220,000 deaths annually in immunocompromised individuals. Munshi et al. explore fungal lipid metabolism in the context of C. neoformans pathogenicity.
INTRODUCTION
Cryptococcus neoformans (C. neoformans) is a pathogenic fungus that presents a leading cause of fungal meningoencephalitis worldwide. Recent reports reveal an annual 278,000 cases of cryptococcal antigenemia, with cryptococcal meningitis being the cause of 15% of AIDS-related deaths (Rajasingham et al., 2017). Naturally occurring cases of cryptococcosis begin by inhalation of fungal spores. Once in the lung, the outcome depends largely on the immune system of the individual. In a situation of suppressed immunity, infection may lead to pneumonia and cryptococcal meningitis. In cases of immunocompetence, these cells are either cleared or may establish a latent infection that will later disseminate upon future immunosuppression.
Once C. neoformans enters the lung, the cells are typically engulfed by an alveolar macrophage in which they can survive and replicate. Similarly, C. neoformans can survive and replicate well in extracellular spaces, such as alveoli, blood, and other tissues. Once engulfed, C. neoformans can move between the phagolysosome and extracellular space without causing harm to the macrophage (Alvarez and Casadevall, 2006; Feldmesser et al., 2000). The intracellular and extracellular environment in the host is distinguished by a prominent difference in pH. Within the phagolysosome, the environment pH is highly acidic, whereas the extracellular environment is typically neutral or slightly alkaline. Adaptation to these starkly contrasting environments is critical for C. neoformans pathogenicity. There is little information regarding how C. neoformans regulates its survival in these two host environments. Previous studies have shown sugar-complexed sphingolipids to be essential for the survival of C. neoformans when grown in media mimicking host acidic or alkaline conditions. Specifically, inositol- or mannose-containing sphingolipids are noted as important for the survival and replication of C. neoformans in conditions similar to the phagolysosome (Luberto et al., 2001). Conversely, glucose-containing sphingolipids have been indicated to be important for survival in conditions mimicking the extracellular environment (Rittershaus et al., 2006). Among sphingolipids, ceramides constitute the simplest class and the basic backbone that precedes other more complex sphingolipids (Aguilera-Romero et al., 2014). Acyl coenzyme A (CoA)-dependent ceramide synthases catalyze the formation of ceramide from a fatty acyl CoA and sphingoid base. Cryptococcal sphingolipids regulate signaling events that lead to the production of virulence factors (Singh and Del Poeta, 2011). Studies in C. albicans (Cheon et al., 2012), S. cerevisiae (Kageyama-Yahara and Riezman, 2006), A. nidulans (Li et al., 2006), and P. pastoris (Ternes et al., 2011) show the presence of two distinct ceramide synthase enzymes.
Although a handful of studies reveal different characteristic functions of ceramide synthases in each eukaryotic species, there is still a lack of concrete evidence for the specific roles of ceramide synthases in the context of sphingolipid biosynthesis of fungi. In this study, we identify and characterize the ceramide synthases of C. neoformans, thereby elucidating several steps in the sphingolipid biosynthetic pathway. We show that the major ceramide synthase of this pathogen is a critical player in its pathogenicity, as well as the hub of numerous cellular functions. We further highlight how a single gene has the capacity to control the dissemination of C. neoformans, therefore suggesting a therapeutic target.
RESULTS
Three Genes in C. neoformans Encode Specific Acyl-CoA-Dependent Ceramide Synthases
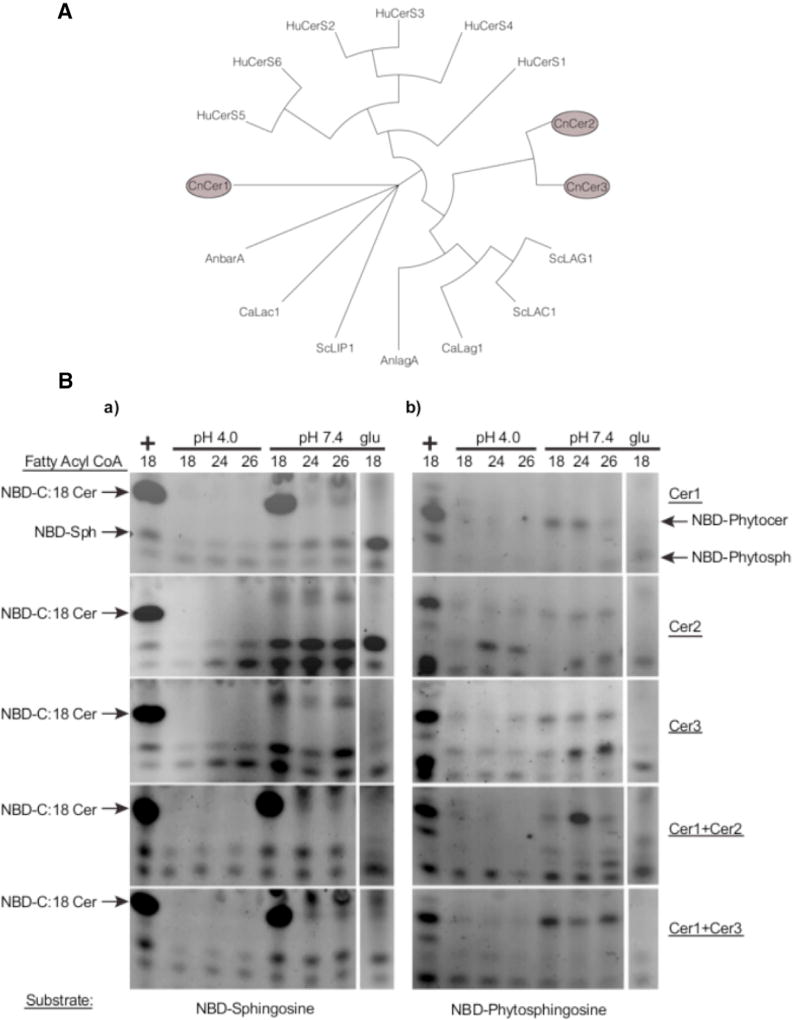
On the basis of evidence of ceramide synthases in other fungi, we performed a bioinformatic search for putative ceramide synthases present in C. neoformans serotype A H99 (WT). Our analysis revealed the presence of three putative ceramide synthases in C. neoformans. The genes CNAG_06717, CNAG_02086, and CNAG_02087 had significant homology to other ceramide synthase genes from A. nidulans, C. albicans, S. cerevisiae, and H. sapiens (Figure 1A). They are referred to in this study as Cer1, Cer2, and Cer3, respectively. A phylogenetic analysis showed that Cer2 and Cer3 have high similarity to each other, as well as to ceramide synthases of S. cerevisiae (ScLac1 and ScLag1) and A. nidulans (AnLagA). The Cer1 amino acid sequence is greatly diverged from Cer2 and Cer3 and has partial similarity to ceramide synthases of C. albicans (CaLag1) and A. nidulans (AnBarA). Reports on these genes show a distinct specificity for synthesis of ceramides used for glucosylceramide (GlcCer) synthesis (Figure 1A) (Li et al., 2006; Rittenour et al., 2011; Cheon et al., 2012). An alignment of the amino acid sequences of these genes revealed conservation of residues that are reportedly important for enzymatic activity (Figure S1D) (Kageyama-Yahara and Riezman, 2006).
Figure 1. Isolation and Characterization of C. neoformans Ceramide Synthases.
(A) Phylogenetic analysis of eukaryotic ceramide synthases. C. neoformans has three ceramide synthases. The entire phylogenetic tree can be roughly divided into three major groups: human cerS genes, the second group containing orthologs of CnCer1, and the third group consisting of S. cerevisiae Lac1 and Lag1, AnLagA, CaLac1, and CnCer2 and CnCer3. ScLip1 is distinct from all of these genes.
(B) Biochemical analysis of C. neoformans ceramide synthases. Thin-layer chromatography showing substrate specificity studies of C. neoformans ceramide synthases. (a) Ceramide synthase assays using NBD-sphingosine as a substrate. Lane 1 (left) is positive control using mammalian Cer1 microsomes. Negative control of each strain grown in2% glucose (right) (see also Figure S3). (b) Ceramide synthase assays using NBD-phytosphingosine as a substrate. Lane 1 (left) is positive control using mammalian Cer1 microsomes. Negative control of each strain grown in 2% glucose (right). Each horizontal panel represents a specific strain.
Ceramide Synthase Cer1 Activity In Vitro Shows Preference for C18 Fatty Acyl CoA
To characterize the activity of each enzyme, we generated strains overexpressing each C. neoformans ceramide synthase in S. cerevisiae using either a 6xHis or 3xHA tag fused protein. These plasmids were transformed in combination into the S. cerevisiae system to generate strains overexpressing both Cer1-Cer2 or Cer1-Cer3. After induction, the proteins were purified by extraction of microsomes (Ternes et al., 2011). As a negative control, these strains were grown without induction (2% glucose) and were used to control for any S. cerevisiae enzyme activity (Figures S3A and S3B).
To characterize the properties of the putative ceramide synthases, an in vitro assay for fungal ceramide synthase was developed (see Experimental Procedures). Using a fluorescent-labeled substrate and fatty acyl CoA, we looked for the formation of NBD-ceramide using microsomal C. neoformans ceramide synthase enzyme by thin-layer chromatography. We observed that the activity of Cer1 is dependent on pH (Figure 1B) and temperature (Figure S3C). The activity of Cer1 was optimal at pH 7.0 and 35°C.
To systematically investigate each enzyme’s specificities, we used combinations of fatty acyl CoAs, with either sphingosine or phytosphingosine at pH 4.0 or 7.0. Cer1 showed a clear preference for C18 fatty acyl CoA and sphingosine as a substrate (Figure 1Ba). When phytosphingosine was used a substrate, we observed production of C18 and C24 phytoceramide in slightly lower amounts (Figure 1Bb). When Cer1 and Cer2 were co-expressed, C24 phytoceramide was the major product. However, when Cer1 and Cer3 were co-expressed, the enzyme activity shifted to C18 and C26 fatty acyl CoA products (Figure 1Bb). These results suggest that the production of ceramide isoforms is regulated by the stoichiometry of the three ceramide synthase proteins of C. neoformans in the presence of different substrate chain lengths. Uninduced cells (2% glucose) showed little to no enzyme activity under these conditions (Figure 1B).
Ceramide Synthases Are Important for the Virulence of C. neoformans
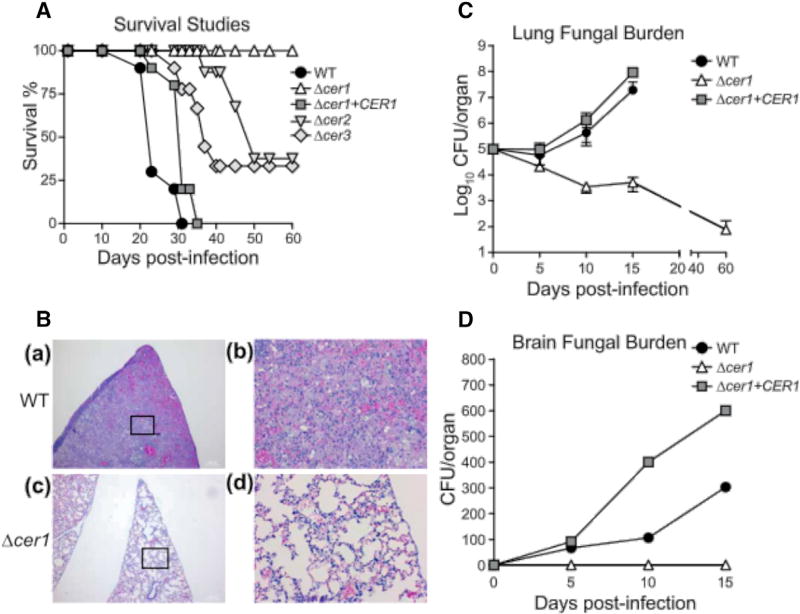
To determine the effect of each ceramide synthase on the virulence of C. neoformans, we created ceramide synthase deletion strains in the WT C. neoformans background (Figures S1A–S1C). Each cell line, containing a deletion cassette or a reintroduced ceramide synthase gene, was tested on CBA/JCrHsd immunocompetent mice. Mice were infected with a normally lethal dose of fungal cells (7 × 105 cells) intranasally to establish cryptococcosis and were monitored for survival. We observed that the average survival of mice infected with WT C. neoformans was 25 ± 6 days, while all mice infected with Δcer1 survived (60 days of observation) (Figure 2A). The average survival of mice infected with the reconstructed gene strain Δcer1+CER1 experienced mortality similar to the WT control, with an average survival of 26 ± 7 days (Figure 2A). Mice infected with Δcer2 and Δcer3 showed a survival pattern distinct from WT (Figure 2A), with each deletion causing ~70% mortality.
Figure 2. Ceramide Synthases Are Important for Virulence of C. neoformans.
(A) Survival studies of CBA/JCrHsd mice infected intranasally with WT, Δcer1, Δcer1 + CER1, Δcer2, and Δcer3. n = 10 mice per group.
(B) Histology of lung tissue for WT(at time of death) and for Δcer1 (day 60). Lung sections were stained with H&E. (a and b) Lung of mice infected with WT. (c and d) Lung of mice infected with Δcer1. Bars, 1,000 µm (a and c), 20 µm (b and d).
(C) Lung tissue burden analysis of WT, Δcer1, and Δcer1+CER1. Data are expressed as mean ± SEM.
(D) Brain fungal burden analysis of WT, Δcer1, and Δcer1+CER1. n = 3 mice at each time point. See also Figure S2.
Tissue burden was assessed throughout the course of the experiment by removal of lungs and brain at days 0, 5, 10, and 15 post-infection. The number of Δcer1 cells in the lung decreased starting at day 5 and was reduced to ~3,500 colony-forming units (CFU)/lung at day 15, showing a decrease in lung CFU of 250-fold. At day 60, we recovered fungal cells in the lung (~500 CFU) but in only two of ten mice, suggesting that most mice cleared the infection (Figure 2C). In contrast to WT C. neoformans infection, Δcer1-infected mice showed no obvious signs of discomfort throughout the course of the experiment and were visually indistinguishable from uninfected mice. The Δcer1 cells were never observed to progress to the brain of infected mice (Figures 2D and S3A). In contrast, a significant number of cells were found in the lungs and brain of mice infected with WT and Δcer1+CER1 strains (Figures 3C and 3D). In both these control experiments, the number of cells in the brain increased over time, demonstrating a normal dissemination and subsequent onset of cryptococcosis. These observations were also confirmed by histopathology of the brain and lung, in which we observed no damage to the lungs and brains of mice infected with Δcer1 (Figure 2B). Mice infected with WT and Δcer1+CER1 showed significant lung and brain tissue damage (Figures 2B and S3).
Figure 3. Sphingolipid Biosynthetic Pathway and Ceramide Species Abundance of C. neoformans Ceramide Synthase Mutants and WT in Distinct Host Conditions In Vitro Using MS Analysis.
(A) Changes in specific lipid classes at pH 4.0.
(B) Changes in specific lipid classes at pH 7.4.
(C and D) Abundance of complex sphingolipid species in WT and ceramide synthase mutants at pH 4.0 (C) and 7.4 (D). All the lipids are normalized to dry lipid weight.
Data are expressed as mean ± SEM. αOH-PCER, α-hydroxy-phytoceramide; CER, ceramide; DHC, dihydroceramide; DHS, dihydrosphingosine; GlcCer, glucosylceramide; IPC, inositolphosphoryl ceramide; PCER, phytoceramide; PHS, phytosphingosine. See also Figure S4 and Tables S2 and S3.
To understand the inflammatory response to C. neoformans infection in the lungs, histopathology was performed at days 1, 3, 5, and 60 (only Δcer1 mice survived to day 60). Lungs of Δcer1-infectedmice showed a high degree of immune cell infiltrate to the lung and alveolar spaces. As the experiment progressed, we observed progressive clearing of the immune cell infiltrate (Figures 2B and S3B). Cells of Δcer1 could be observed in several areas of the lung until day 5. These cells appeared to have difficulty completing replication and had a pseudohyphal morphology. Lungs infected with WT showed a persistently high level of immune cell infiltrate throughout the time course, enlarged C. neoformans capsules, and fungal pneumonia, which was not observed for Δcer1 (Figure S2B). The total burden of WT C. neoformans cells increased throughout the time course (Figure 2).
Identification of Lipid Changes on Ceramide Synthase Deletion
The sphingolipid pathway in C. neoformans can be separated into two major branches: substrates that lead to the generation of glucose-containing sphingolipids such as GlcCer and those that lead to inositol-containing sphingolipids such as inositol phosphorylceramide (IPC). To assess the specific roles of each gene in lipid metabolism, we analyzed each knockout strain with lipidomic-focused mass spectrometry. The analysis was performed on cells grown in in vitro conditions (5% CO2, 37°C, DMEM) mimicking host-like growth conditions.
Because fungal cells produce a diverse array of ceramide species, we developed mass spectrometry detection protocols for an array of the most abundant ceramide lipids. When exposed to an acidic environment, WT C. neoformans showed accumulation of dihydrosphingosine as well as higher total biomass of GlcCer species (Figure 3). We also observed higher abundance of phytosphingosine and certain long-chain α-OH phytoceramides (C24, C26). This increase was propagated downstream in the pathway by increased levels of 42:0:4, 42:0:5, and 44:0:4 chain length IPCs (Figures 3 and S4). In contrast, at alkaline pH, sphingosine-1-phosphate, dihydrosphingosine-1-phosphate, and phytosphingosine-1-phosphate were twice as abundant as those in acidic pH (Figure S4). Therefore, the data show not only that C. neoformans changes the metabolism of several lipid species when it is exposed to different host conditions but, more important, that this change depends on specific ceramide isoforms to generate sufficient amounts of the resulting complex sphingolipids. The lipid profile of Δcer1 was significantly perturbed from WT compared with the milder phenotypes of Δcer2 and Δcer3. We observed that Cer1 was the major ceramide synthase responsible for using C18 fatty acyl CoA to generate C18 ceramides (Figures 1B and 3). As C18 ceramides are required to synthesize aOH-C19:2/C18 GlcCer, the most abundant GlcCer in either condition, the strain Δcer1 is lacking the vast majority of its normal glucose-containing complex sphingolipids (Figure 3C). C24 and C26 lipids were not significantly depleted in the Δcer1 strain but also represented a tiny fraction of total GlcCer abundance in the WT strain (Figure 3C). This observation agrees with the in vitro findings of chain-length specificity for Cer1 (Figure 1B). In the Δcer1 strain, depletion of IPCs generated from non-hydroxylated and α-hydroxylated C18 phytoceramides contrasts with the accumulation of long-chain IPCs. Notably, Δcer1 and Δcer3 showed a significant depletion of C26 phytoceramides in acidic conditions (Figure 3A).
These data indicate that upon loss of Cer1, Cer2 and Cer3 can partially compensate for production of certain sphingolipid isoforms but are insufficient for production of the major C18 ceramide isoforms. Broadly speaking, of the lipid isoforms showing significant change, these changes were most pronounced under acidic conditions (Figures 3 and S5). The levels of the major IPC, IPC 42:0:4, remained relatively unchanged in the deletion strains. Δcer2 showed little to no depletion of lipids but a marked increase in dihydrosphingosine, C18 dihydroceramide, and total short-chain GlcCers. Additionally, Δcer2 showed a clear abundance of most C18 lipids in the IPC pathway: phytosphingosine, phytoceramides, αOH phytoceramides, and C36 IPCs all were significantly more abundant compared with WT. Meanwhile, Δcer3 showed a decrease in lipids along the GlcCer branch of the pathway as well as a decrease in C24 and C26 lipids in the IPC pathway under alkaline conditions. Together, these data suggest that Cer1 is critical for the biosynthesis of C18 ceramides, where Δcer2 and Δcer3 show more subtle lipid phenotypes and appear to fine-tune the abundance of less common lipids (Figure S4; Data S1).
Effect of Ceramide Synthase on the In Vitro Growth of C. neoformans
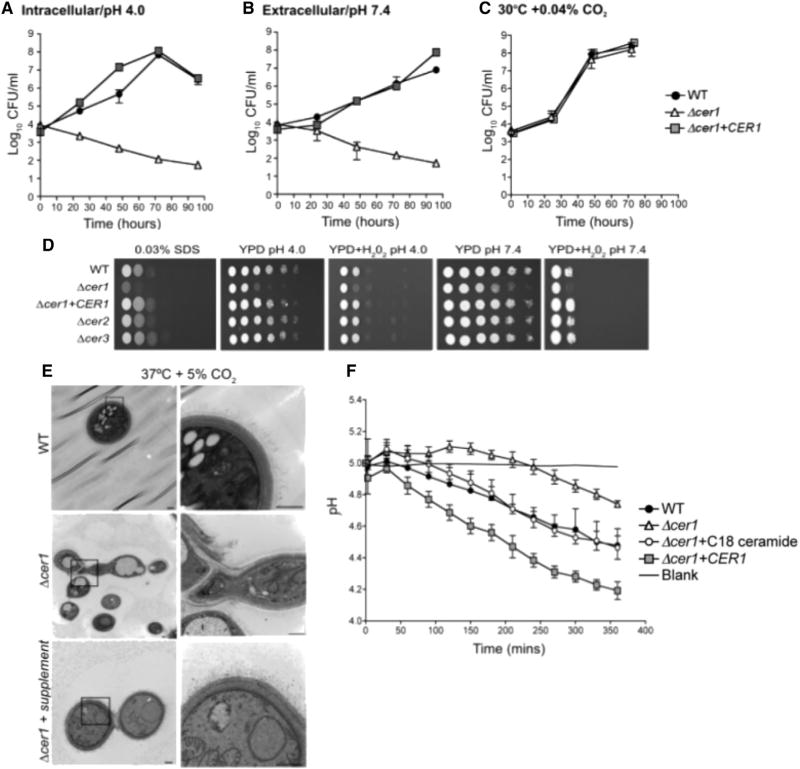
To evaluate the effect of gene deletion in in vitro cell culture growth, we performed growth assays of C. neoformans WT, Δcer1, and Δcer1+CER1 strains. Growth was analyzed in conditions mimicking host intracellular and extracellular conditions, using DMEM at neutral/alkaline or acidic conditions at 37°C and 5% CO2. As shown in Figures 4A and 4B, Δcer1 cells showed a distinct lack of growth at both conditions mimicking host environments. Δcer1 viability began to decrease at 24 hr in acidic as well as alkaline conditions. Loss of viability was not observed at 30°C and atmospheric level of CO2 (Figure 4C). These results indicate the deletion of Δcer1 is important for the survival and proliferation of C. neoformans in host intracellular and extracellular conditions.
Figure 4. Phenotypic Analysis of Ceramide Synthase Mutants.
(A) In vitro growth of WT, Δcer1, and Δcer1+CER1 at 37°C, 5% CO2, pH 4.0 (intracellular).
(B) In vitro growth of WT, Δcer1, and Δcer1+CER1 at 37°C, 5% CO2, pH 7.4 (extracellular).
(C) In vitro growth of WT and Δcer1 in YPD, 30°C, 0.04% CO2.
(D) Serial dilutions of WT, Δcer1, Δcer1 +CER1, Δcer2, and Δcer3 on solid YPD media supplemented with SDS or H2O2.
(E) Transmission electron microscopy of WT, Δcer1, and Δcer1 supplemented with ceramide mixture (Matreya). Bar, 500 nm.
(F) Pma1 proton pump activity of WT, Δcer1, Δcer1+CER1, and Δcer1 + C18 ceramide (Avanti Polar Lipids) measured by glucose-dependent medium acidification. Data are expressed as mean ± SEM.
For (A)–(C), data are expressed as mean ± SEM.
To determine the effect of cell wall stress on the deletion strains, a spot assay was performed by exposing WT, Δcer1, and Δcer1+CER1 to 0.03% SDS. We observed that Δcer1 was more sensitive to cell wall stress compared with WT and Δcer1+CER1 (Figure 4D). We tested the resilience of these strains to oxidative stress by exposing dilutions of these cells to hydrogen peroxide at pH 4 and 7.4 (Figure 4D). Δcer1 was hypersensitive to oxidative stress at both pH values. The hypersensitivity of Δcer1 to low pH and other stresses are significant considering that the spot assay was done using rich medium (YPD) because it could not be performed in minimum medium for the growing defect phenotype of the mutant.
Phenotype Analysis of Ceramide Synthase Mutant Strains
To assess the physiological effects of the deletion of ceramide synthase genes, we analyzed the phenotypes of these cells under several conditions with a focus on alteration of virulence factors. The deletion of Cer1, but not Cer2 or Cer3, generated defects in cell morphology. Capsule visualization by India ink staining revealed that cells of Δcer1 had cell division defects leading to the development of elongated cells with a smaller capsule (Figure S5B). When Δcer1 was grown in rich medium (YPD), the cells showed normal morphology, with a few cells showing multiple enlarged buds that remained attached. However, upon transfer to host-like growth conditions (5% CO2, 37 ° C, DMEM), Δcer1 cells showed gross morphological defects and inability to complete cytokinesis. A lifespan study of these cells showed that the average replicative lifespan (RLS) of Δcer1 was only 6.5 generations, whereas those of WT and Δcer1+CER1 were 27 and 30 generations respectively (Figure S5A). For Δcer1, after 6.5 ± 2 generations, the cells formed elongated pseudohyphal-like structures in which new buds were impossible to separate from the mother cell. Despite the inability to separate, the cells continued to elongate for several hours.
We performed transmission electron microscopy to further observe these changes in cellular structure. Δcer1 cells showed detachment of the cell wall from the plasma membrane in many cells. The elongated ‘‘hyphal-like’’ structures observed in histological and India ink staining were found to be daughter cells that were unable to complete cytokinesis. Interestingly, we also observed that the cell wall structure of Δcer1 looked very different from that of WT. Although the WT cells showed well-defined, distinct layers resulting in a compact cell wall, the cell wall of Δcer1 appeared less compact, with no clear separation between cell wall layers (Figure 4E). The fibrillar structures forming the polysaccharide capsule were smaller and less distinct than that of WT (Figure 4E). We hypothesize that the layers of the cell wall of Δcer1 are inhibitory to cytokinesis and cell separation under two conditions: when the cells are exposed to host-like conditions and when cells are grown in the absence of rich, ceramide lipid-containing media. To confirm the role of ceramides in the formation of such a drastic cellular defect, the cells of Δcer1 were supplemented with a cocktail of natural ceramides (Matreya). Upon supplementation, we observed that many of the previously observed cellular defects were recovered (Figure 4E). This cocktail predominantly contains a mixture of C18 and C24 hydroxylated and non-hydroxylated ceramides. Together these observations suggest that ceramides synthesized by Cer1 are critical for proper cytokinesis in stressful host conditions, in which increased cell wall thickness introduces a hindrance for proper daughter cell budding.
C18 Ceramides Generated by Cer1 Are Important for Acidic Tolerance of C. neoformans
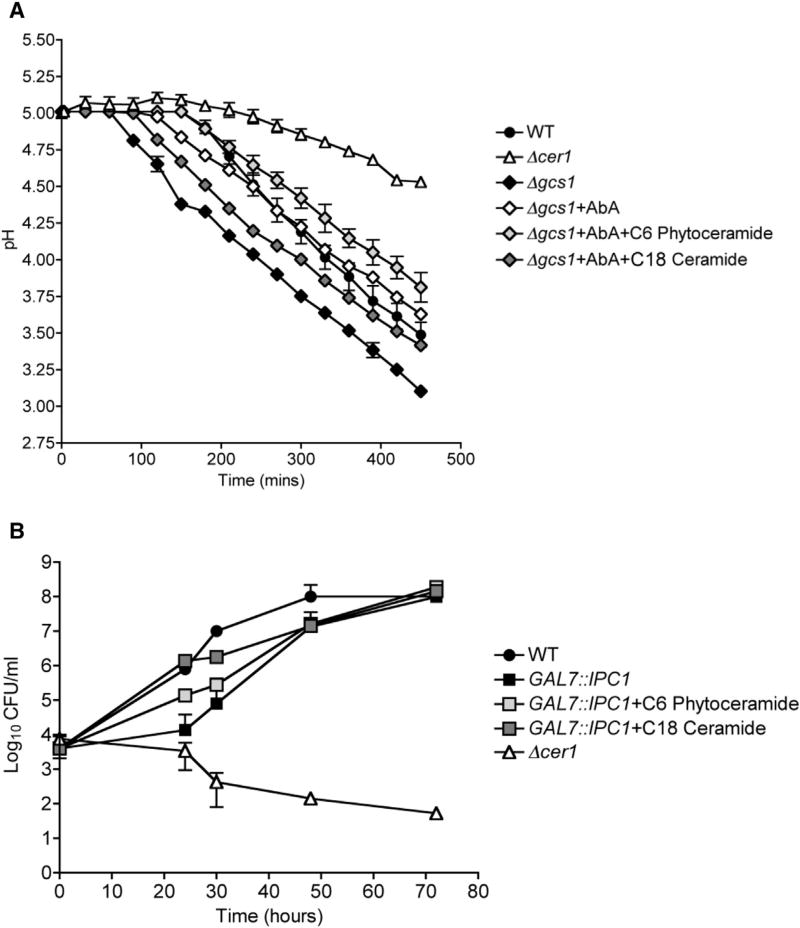
We checked the efficiency of the plasma membrane proton pump, Pma1, upon deletion of a ceramide synthase in C. neoformans. Pma1 is a crucial mediator of C. neoformans virulence, as the proton pump regulates C. neoformans cytosolic pH. This is a particularly important function for C. neoformans growth inside acidic macrophage lysosomes. A colorimetric pH indicating dye was used to measure glucose-dependent proton efflux over a time course (Soteropoulos et al., 2000). Our results showed that Δcer1 cells acidify medium at a much slower rate than WT and Δcer1+CER1 strains (Figure 4F). We observed an increase in Pma1 efficiency in Δcer1 cells upon supplementation with C18 ceramide (Figure 4F). To further confirm to the role of ceramides in the observed phenotypes, we tested Pma1 activity of Δgcs1 strain, as well as the Δgcs1 strain treated with an IPC1 inhibitory drug, aureobasidin A (AbA). We observed that Δgcs1 had high Pma1 activity, which was reduced upon treatment with AbA (Figure 5A). Interestingly, supplementation with C18 ceramides, but not phytoceramides, increased Pma1 activity of AbA-treated Δgcs1 cells (Figure 5A). To ensure that the Pma1 activity data were not attributable to higher or lower rates of cell death, we measured cell viability at the end of each experiment and found no difference between strains (data not shown). To further clarify the role of intermediate ceramides, we used a genetically downregulated IPC1 strain (GAL7::IPC1) that has been reported to show reduced growth in acidic conditions. The growth defect of this strain was compensated by supplementation C18 ceramide and to a smaller extent by C6 phytoceramide (Figure 5B). These data indicate that the intermediate ceramide isoforms, not the terminal IPC and GlcCer isoforms, play a major role in regulation of Pma1 and normal cell growth.
Figure 5. Effect of Ceramide on Pma1 Activity and on Cell Growth.
(A) Pma1 proton pump activity of WT, Δcer1, Δgcs1, Δgcs1+AbA, Δgcs1+AbA+C6 phytoceramide, and Δgcs1+AbA+C18 ceramide measured by glucose-dependent medium acidification. Data are expressed as mean ± SEM.
(B) In vitro growth of WT, GAL7::IPC1, GAL7:: IPC1+C6 phytoceramide, GAL7::IPC1+ C18 ceramide, and Δcer1 at 37°C, 5% CO2, pH 4.0. Data are expressed as mean ± SEM.
DISCUSSION
An understanding of the mechanisms driving C. neoformans pathogenicity is an increasingly important area of research because of the growing number of cases of cryptococcosis worldwide. Deeper insight into how this pathogen efficiently maintains itself within immunocompromised hosts will bring us to closer to finding a way to control its growth and dissemination. Dynamic adjustment of the sphingolipid profile in C. neoformans cells has been reported to play a significant role in C. neoformans’s ability to grow in either highly acidified phagocytic environment or slightly alkaline blood, alveolar, and brain tissue environments. Previous studies attempting to elucidate this pathway have uncovered important pieces of this puzzle (Del Poeta et al., 2015; Rittershaus et al., 2006; Luberto et al., 2001). In the present study, we provide a more comprehensive view of the involved genes and show their importance in the lipid profile switch between these two contrasting environments.
Here, we show that ceramide synthase Cer1 is a major factor in the pathogenicity of C. neoformans. We observed that deletion of each ceramide synthase showed a survival curve that was significantly divergent from WT infection, with Δcer1 being completely avirulent with 80% of mice totally clearing the infection within 60 days. Lack of in vitro growth in host intracellular and extracellular conditions suggests that Δcer1 lacks the ability to efficiently thrive in the host upon infection. In stark contrast to previous studies on Δgcs1 and GAL7::IPC1, cells of Δcer1 quickly begin to die when grown in minimal medium conditions regardless of either pH. An inhibitor of Cer1 could potentially be a very good drug for the treatment of cryptococcosis and other fungal diseases.
There are studies that suggest a role of phytoceramides in the activity of plasma membrane proton pumping H+-ATPase, Pma1 (Farnoud et al., 2014; Keniya et al., 2013; Soteropoulos et al., 2000). Our results show that Δcer1 has a marked reduction in the activity of Pma1, thereby suggesting a role, possibly through an indirect mechanism, of C18 ceramides in the ability of C. neoformans to maintain its survival within the highly acidic phagolysosome. Increased Pma1 activity in the Δgcs1 mutant suggests two possible models of ceramide species mediated regulation of Pma1: it is possible that the lack of GlcCer species in Δgcs1 relieves an inhibitory pressure on Pma1 activity, causing the observed increase. Alternatively, a buildup of intermediate ceramide compounds or IPCs in response to a lack of Gcs1 (Table S2) could have a positive effect on Pma1 activity, which could also result in the observed increase. Pma1 activity in the presence of AbA and Δgcs1 indicates intermediate ceramide compounds play a major role in the enzyme’s activity.
Δcer1 cells have critical defects in cytokinesis in the presence of host-like environmental stresses. The cell wall is improperly anchored to the plasma membrane in many cells; consequently the membrane and cell wall structure may be inhibiting daughter cell separation. It remains unclear if the altered morphology of the cell wall or the lack of cell wall adherence to the plasma membrane is the major contributor to this phenotype. The presence of irregular amounts of long-chain ceramides and complex sphingolipids could be affecting the rigidity of the cell wall and/or plasma membrane.
We observed that each knockout strain had distinct alterations of ceramides and downstream lipids, as shown by lipidomic analysis, indicating that the genes are not functionally redundant, and each likely plays a specific role in maintaining the optimum lipid profile for C. neoformans membranes. Despite numerous attempts to create a double-mutant Δcer2Δcer3, we have been unsuccessful. We suspect, but are unable to confirm because of tight genetic linkage, that the presence of either Cer2 or Cer3 is essential to cell survival. Furthermore, the inability of Δcer1 cells to survive in acidic conditions, despite abundant amounts of IPC-42:0:4, strongly suggests an important role of C18 ceramides for this phenotype.
In summary, this study is a novel insight into the critical importance of ceramides and ceramide synthases in cryptococcal pathogenicity. Cer1 predominately uses C18 fatty acid chain substrates in order to synthesize specific ceramides and complex sphingolipids that are of crucial importance toward C. neoformans pathogenicity. Our results can be extrapolated to several other pathogenic fungi and therefore provide new hope for immunocompromised individuals susceptible to fungal infections.
EXPERIMENTAL PROCEDURES
Strains, Plasmids, and Media
The strains used in this study are C. neoformans var. grubii strain H99 as wild-type (WT) and S. cerevisiae BY4741. The bacterial strain used was Escherichia coli DH5-α Max Efficiency (Invitrogen, Carlsbad, CA, USA) as competent cells. The three putative ceramide synthase genes in this study have the following identifiers: CNAG_06717 (GenBank: XM_012192296), CNAG_02086 (GenBank: XM_012194542), and CNAG_02087 (GenBank: XM_012194543). pYES2/CT was used for overexpression of CNAG_06717 in S. cerevisiae BY4741 and pRS425 for CNAG_02086 and CNAG_02087. C C. neoformans strains were routinely grown in YPD broth at 30°C and 0.04% CO2 shaking overnight. DMEM buffered with 25 mM HEPES (pH 4.0 or 7.4) was used to grow C. neoformans at 37°C in the presence of 5% CO2. S. cerevisiae transformed with pYES2/CT was grown in YNB (ura−), and S. cerevisiae transformed with pRS425 was grown in synthetic leucine (leu−) dropout media. Strains containing both vectors were grown in synthetic leu−ura− dropout media. Bacterial strains were grown at 37°C in Luria-Bertani media containing 75 mg/L of ampicillin (Sigma-Aldrich). All primers are specified in Table S1.
Phylogenetic Analysis of Putative Ceramide Synthase Genes
Exact maximum likelihood phlyogenetic tree construction was performed using TreePuzzle (Schmidt et al., 2002; Schmidt and Von Haeseler, 2007) with 1,000 quartet puzzling steps. Dendroscope was used to visualize the resulting phylogenetic tree (Huson et al., 2007; Huson and Scornavacca, 2012).
Fluorescent Cryptococcal Ceramide Synthase Assay
An assay for ceramide synthase activity was adapted (Kim et al., 2012; Tidhar et al., 2015) with minor changes. Enzymes were extracted as microsomes (see Supplemental Experimental Procedures). Reactions contain 10 µM NBD-sphingosine or NBD-phytosphingosine (Avanti Polar Lipids) and 50 µM fatty acyl CoA (C18, C24, C26) (Avanti Polar Lipids, Alabaster, AL, USA). One hundred fifty micrograms of microsomal protein was added per reaction. Reactions were incubated at 35°C for 90 min, followed by enzyme inactivation with 2:1 chloroform/methanol. The lipids were extracted and dried in a SpeedVac (SPD 2010; Thermo Fisher Scientific) and resuspended in 100% methanol. The reaction was analyzed using thin-layer chromatography, using chloroform/methanol/water (8:1:0.1, v/v/v) as the solvent mixture.
Virulence Studies and Histology Analysis in a Murine Mouse Model of Cryptococcosis
Three- to 4-week-old female CBA/JCrHsd mice (Harlan Laboratories) were used for all experiments. Mice were infected with WT, Δcer1, Δcer2, Δcer3, and Δcer1+CER1 at a concentration of 3.5×107 cells/mL. For survival studies, ten CBA/JCrHsd mice per strain were infected intranasally with 7 × 105 cells/20 µL. For tissue burden analysis, nine mice per strain were used. Organs were excised and homogenized in 10 mL PBS using Stomacher 80 (Seward) for 2 min at high speed. For histopathology analysis, three mice per experimental group were used. Mice organs were fixed in 3.7% formaldehyde in paraffin and stained with H&E and mucicarmine. All animal procedures were approved by the Stony Brook University Institutional Animal Care and Use Committee and followed the guidelines of American Veterinary Medical Association.
Extraction and Mass Spectrometry Analysis of Yeast Sphingolipids
Lipid extraction was performed according to the methods of Bligh and Dyer (1959). After measuring the dry weights, the samples were subject to base hydrolysis (Clarke and Dawson, 1981). The extracts were dried in a SpeedVac (SPD 2010, ThermoFisher Scientific, Waltham, MA, USA). All internal standards were added prior to lipid extraction. Mass spectrometry (MS) analysis detailed in Supplemental Experimental Procedures.
In Vitro Growth Studies
Cells of C. neoformans WT, Δcer1, and Δcer1+CER1 were grown in 10 mL DMEM (buffered with HEPES [pH 4.0 or 7.4]) to a final density of 104 cells/mL and incubated in shaker incubator at 37°C with 5% CO2 or YPD (30°C, 0.04% CO2) where mentioned. GAL7::IPC1 was grown as previously described (Luberto et al., 2001). Aliquots were taken at the time points indicated. For cell wall stability, cells were spotted in serial dilutions on YPD plates with 0.05% SDS. For oxidative stress, cells were spotted on YPD containing 2 mM H2O2. C6 phytoceramide (50 µM) (Matreya) and C18 ceramide (Avanti Polar Lipids) were supplemented where mentioned.
Transmission Electron Microscopy
C. neoformans strains were grown for 24 hr in DMEM (pH 4.0 or 7.4) at 37°C + 5% CO2. Cells were later washed and fixed in 3% EM-grade glutaraldehyde solution for 2 hr. Cells were supplemented with 50 µM ceramide mix where indicated (Matreya). (Additional sample preparation details are provided in Supplemental Experimental Procedures.) Visualization of samples was performed using a FEI TeCnai12 BioTwinG2 transmission electron microscope. Digital images were acquired with an AMT XR-60 charge-coupled device (CCD) digital camera system.
Glucose-Dependent Medium Acidification to Measure Plasma Membrane H+-ATPase
Glucose-dependent medium acidification was monitored by a modification of a procedure described previously (Perlin et al., 1988; Soteropoulos et al., 2000). Briefly, cells of WT, Δcer1, Δcer1+CER1, and Δgcs1 were grown to mid-log phase in YPD. The following day, cells were transferred to DMEM at pH 4.0 for 24 hr. AbA (0.05 µg/mL) and 50 µM ceramide supplements were added where mentioned. Cells were then harvested, washed, and resuspended in 100 mM KCl (pH 5.0), shaking at 30°C for 1 hr. These samples were then stored at 4°C overnight prior to use. For the assay, cells were concentrated to a final A590 of approximately 2.0. Twenty microliters of cells along with 155 µL of bromophenol blue (50 µg/mL) in 100 mM KCl (pH 5.0) and 20 µL 20% (w/v) glucose was added to initiate the reaction. Medium acidification was monitored at 590 nm over a period of 5 hr (data point every 3 mins) in a microplate reader (SpectraMax M5)
Statistics
Data are expressed as mean ± SEM. Calculations were made and data graphed using GraphPad Prism software version 6.0.
Supplementary Material
Highlights.
Characterization of Cryptococcus neoformans (C. neoformans) ceramide synthases
A virulence role for ceramides in C. neoformans infection
Ceramide pathway affects Pma1 activity
Cer1 primarily produces ceramides with chain lengths of 18 carbons
Acknowledgments
This work was supported by NIH grants AI116420 and AI125770 to M.D.P. and AI127704 to B.C.F. and in part by National Cancer Institute (NCI) grant P01-CA97132 to C.L. for project #4. M.D.P. is a Burroughs Welcome Investigator in Infectious Diseases. We are grateful to Dr. Martha Furie, Dr. Jamie Konopka, Dr. Luis Martinez, Dr. Amir Farnoud, Dr. Visesato Mor, Dr. Aaron Neiman, and members of the Del Poeta and Luberto labs for valuable inputs. Special thanks to Dr. Can Senkal for providing mammalian CER1 microsomes. We thank Susan Van Horn and the transmission electron microscopy (TEM) facility at the Central Microscopy Imaging Center (C-MIC) at Stony Brook University. We thank Yelena Altschuller for help with plasmid construction. The Stony Brook Proteomics facility, Research Histology core, and the McClain lab provided valuable assistance for several experimental procedures. We thank Jasna Boudard and Zoe Reifsnyder for their help with graphics and figures.
Footnotes
Supplemental Information includes Supplemental Experimental Procedures, five figures, three tables, and one data file and can be found with this article online at https://doi.org/10.1016/j.celrep.2018.01.035.
AUTHOR CONTRIBUTIONS
Conceptualization, M.A.M., J.M.G., C.L., and M.D.P.; Methodology, M.A.M. and J.M.G.; Investigation, M.A.M.; Lipidomics, A.S. and R.R.; Writing – Original Draft, M.M. and J.M.G.; Writing – Review & Editing, J.M.G. and M.D.P.; Replicative Lifespan Study, T.B. and B.C.F.; Funding Acquisition, Resources, and Supervision, M.D.P.
DECLARATION OF INTERESTS
M.D.P. is a co-founder and chief scientific officer (CSO) of MicroRid Technologies. All other authors have no conflict of interest.
References
- Aguilera-Romero A, Gehin C, Riezman H. Sphingolipid homeostasis in the web of metabolic routes. Biochim. Biophys. Acta. 2014;1841:647–656. doi: 10.1016/j.bbalip.2013.10.014. [DOI] [PubMed] [Google Scholar]
- Alvarez M, Casadevall A. Phagosome extrusion and host-cell survival after Cryptococcus neoformans phagocytosis by macrophages. Curr. Biol. 2006;16:2161–2165. doi: 10.1016/j.cub.2006.09.061. [DOI] [PubMed] [Google Scholar]
- Bligh EG, Dyer WJ. A rapid method of total lipid extraction and purification. Can. J. Biochem. Physiol. 1959;37:911–917. doi: 10.1139/o59-099. [DOI] [PubMed] [Google Scholar]
- Cheon SA, Bal J, Song Y, Hwang HM, Kim AR, Kang WK, Kang HA, Hannibal-Bach HK, Knudsen J, Ejsing CS, Kim JY. Distinct roles of two ceramide synthases, CaLag1p and CaLac1p, in the morphogenesis of Candida albicans. Mol. Microbiol. 2012;83:728–745. doi: 10.1111/j.1365-2958.2011.07961.x. [DOI] [PubMed] [Google Scholar]
- Clarke NG, Dawson RM. Alkaline O leads to N-transacylation. A new method for the quantitative deacylation of phospholipids. Biochem. J. 1981;195:301–306. doi: 10.1042/bj1950301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Del Poeta M, Nimrichter L, Rodrigues ML, Luberto C. Correction: synthesis and biological properties of fungal glucosylceramide. PLoS Pathog. 2015;11:e1004886. doi: 10.1371/journal.ppat.1004886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farnoud AM, Mor V, Singh A, Del Poeta M. Inositol phosphosphingolipid phospholipase C1 regulates plasma membrane ATPase (Pma1) stability in Cryptococcus neoformans. FEBS Lett. 2014;588:3932–3938. doi: 10.1016/j.febslet.2014.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feldmesser M, Kress Y, Novikoff P, Casadevall A. Cryptococcus neoformans is a facultative intracellular pathogen in murine pulmonary infection. Infect. Immun. 2000;68:4225–4237. doi: 10.1128/iai.68.7.4225-4237.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huson DH, Scornavacca C. Dendroscope 3: an interactive tool for rooted phylogenetic trees and networks. Syst. Biol. 2012;61:1061–1067. doi: 10.1093/sysbio/sys062. [DOI] [PubMed] [Google Scholar]
- Huson DH, Richter DC, Rausch C, Dezulian T, Franz M, Rupp R. Dendroscope: an interactive viewer for large phylogenetic trees. BMC Bioinformatics. 2007;8:460. doi: 10.1186/1471-2105-8-460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kageyama-Yahara N, Riezman H. Transmembrane topology of ceramide synthase in yeast. Biochem. J. 2006;398:585–593. doi: 10.1042/BJ20060697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keniya MV, Cannon RD, Nguyễn Â, Tyndall JD, Monk BC. Heterologous expression of Candida albicans Pma1p in Saccharomyces cerevisiae. FEMS Yeast Res. 2013;13:302–311. doi: 10.1111/1567-1364.12035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim HJ, Qiao Q, Toop HD, Morris JC, Don AS. A fluorescent assay for ceramide synthase activity. J. Lipid Res. 2012;53:1701–1707. doi: 10.1194/jlr.D025627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Du L, Yuen G, Harris SD. Distinct ceramide synthases regulate polarized growth in the filamentous fungus Aspergillus nidulans. Mol. Biol. Cell. 2006;17:1218–1227. doi: 10.1091/mbc.E05-06-0533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luberto C, Toffaletti DL, Wills EA, Tucker SC, Casadevall A, Perfect JR, Hannun YA, Del Poeta M. Roles for inositol-phosphoryl ceramide synthase 1 (IPC1) in pathogenesis of C. neoformans. Genes Dev. 2001;15:201–212. doi: 10.1101/gad.856001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perlin DS, Brown CL, Haber JE. Membrane potential defect in hygromycin B-resistant pma1 mutants of Saccharomyces cerevisiae. J. Biol. Chem. 1988;263:18118–18122. [PubMed] [Google Scholar]
- Rajasingham R, Smith RM, Park BJ, Jarvis JN, Govender NP, Chiller TM, Denning DW, Loyse A, Boulware DR. Global burden of disease of HIV-associated cryptococcal meningitis: an updated analysis. Lancet Infect. Dis. 2017;17:873–881. doi: 10.1016/S1473-3099(17)30243-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rittenour WR, Chen M, Cahoon EB, Harris SD. Control of glucosylceramide production and morphogenesis by the Bar1 ceramide synthase in Fusarium graminearum. PLoS ONE. 2011;6:e19385. doi: 10.1371/journal.pone.0019385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rittershaus PC, Kechichian TB, Allegood JC, Merrill AH, Jr, Hennig M, Luberto C, Del Poeta M. Glucosylceramide synthase is an essential regulator of pathogenicity of Cryptococcus neoformans. J. Clin. Invest. 2006;116:1651–1659. doi: 10.1172/JCI27890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt HA, Von Haeseler A. Maximum-likelihood analysis using TREE-PUZZLE. Curr. Protoc. Bioinformatics. 2007;Chapter 6(Unit 6.6) doi: 10.1002/0471250953.bi0606s17. [DOI] [PubMed] [Google Scholar]
- Schmidt HA, Strimmer K, Vingron M, von Haeseler A. TREE-PUZZLE: maximum likelihood phylogenetic analysis using quartets and parallel computing. Bioinformatics. 2002;18:502–504. doi: 10.1093/bioinformatics/18.3.502. [DOI] [PubMed] [Google Scholar]
- Singh A, Del Poeta M. Lipid signalling in pathogenic fungi. Cell. Microbiol. 2011;13:177–185. doi: 10.1111/j.1462-5822.2010.01550.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soteropoulos P, Vaz T, Santangelo R, Paderu P, Huang DY, Tamás MJ, Perlin DS. Molecular characterization of the plasma membrane H(+)-ATPase, an antifungal target in Cryptococcus neoformans. Antimicrob. Agents Chemother. 2000;44:2349–2355. doi: 10.1128/aac.44.9.2349-2355.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ternes P, Wobbe T, Schwarz M, Albrecht S, Feussner K, Riezman I, Cregg JM, Heinz E, Riezman H, Feussner I, Warnecke D. Two pathways of sphingolipid biosynthesis are separated in the yeast Pichia pastoris. J. Biol. Chem. 2011;286:11401–11414. doi: 10.1074/jbc.M110.193094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tidhar R, Sims K, Rosenfeld-Gur E, Shaw W, Futerman AH. A rapid ceramide synthase activity using NBD-sphinganine and solid phase extraction. J. Lipid Res. 2015;56:193–199. doi: 10.1194/jlr.D052001. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.