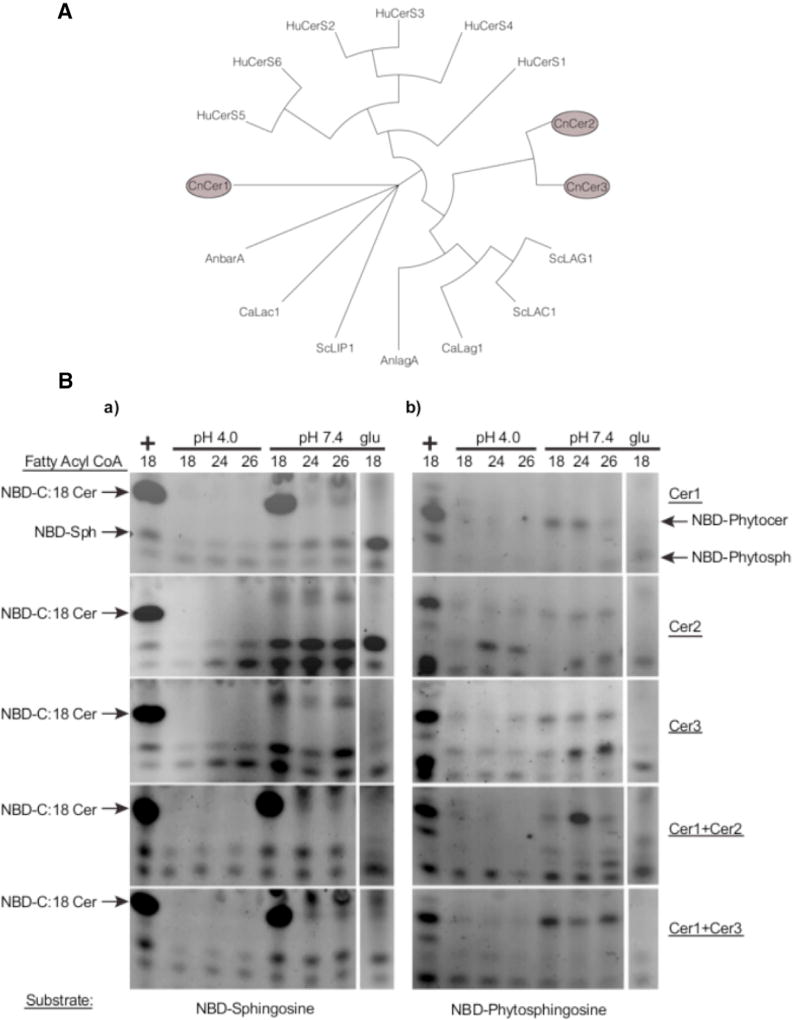
Figure 1. Isolation and Characterization of C. neoformans Ceramide Synthases.
(A) Phylogenetic analysis of eukaryotic ceramide synthases. C. neoformans has three ceramide synthases. The entire phylogenetic tree can be roughly divided into three major groups: human cerS genes, the second group containing orthologs of CnCer1, and the third group consisting of S. cerevisiae Lac1 and Lag1, AnLagA, CaLac1, and CnCer2 and CnCer3. ScLip1 is distinct from all of these genes.
(B) Biochemical analysis of C. neoformans ceramide synthases. Thin-layer chromatography showing substrate specificity studies of C. neoformans ceramide synthases. (a) Ceramide synthase assays using NBD-sphingosine as a substrate. Lane 1 (left) is positive control using mammalian Cer1 microsomes. Negative control of each strain grown in2% glucose (right) (see also Figure S3). (b) Ceramide synthase assays using NBD-phytosphingosine as a substrate. Lane 1 (left) is positive control using mammalian Cer1 microsomes. Negative control of each strain grown in 2% glucose (right). Each horizontal panel represents a specific strain.