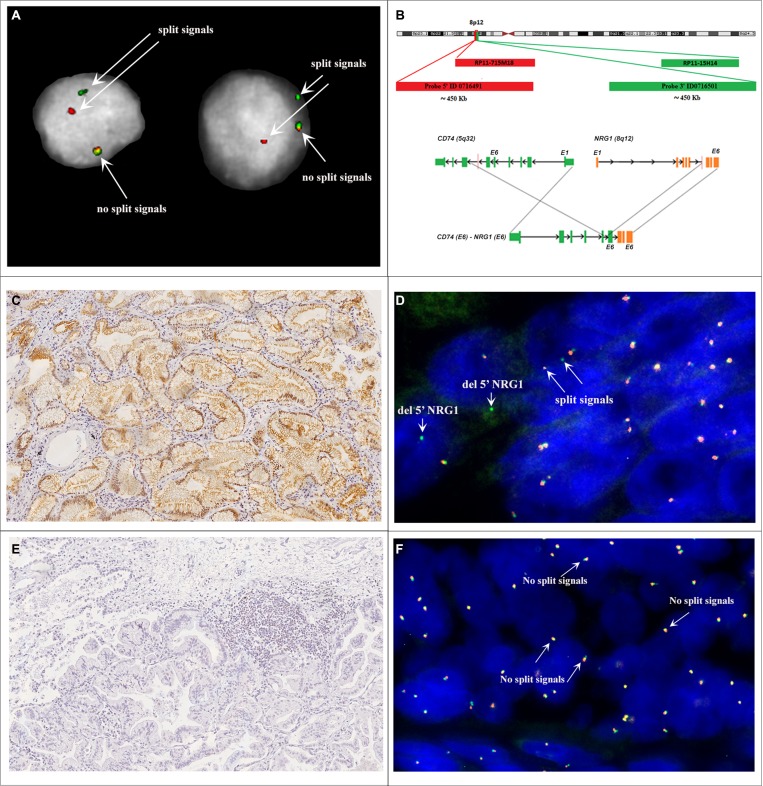
Figure 2.
(A) Nuclei MD-175. (B) Schematic illustration of FISH probes map used for detection of NRG1 rearrangement. For break-apart FISH assay telomeric BAC RP11-715M18 (chr8:32,264,921-32,433,449) and centromeric RP11-15H14 (chr8:32,679,447-32,840,717) were co-hybridized. Further FISH tests were performed by use of custom 5′-Red ID 0716491 (∼450 Kb) and 3’-Green ID 0716501 (∼450 Kb) probes. NRG1 gene, BACs and custom probes are represented not in scale. (C) IMA (case MD-55) showing p-ErbB3 immunoreactivity. Magnification X20. (D) FISH analysis of case MD-55 shows disrupted NRG1 locus. Red arrows highlight the split of the green and red probe signals (hybridizing to 3′ and 5′ regions of NRG1 respectively). (E) IMA (case LCCH-556) without p-ErbB3 immunoreactivity. Magnification X20. (F) FISH analysis of case LCCH-556 shows the wild-type NRG1 locus.