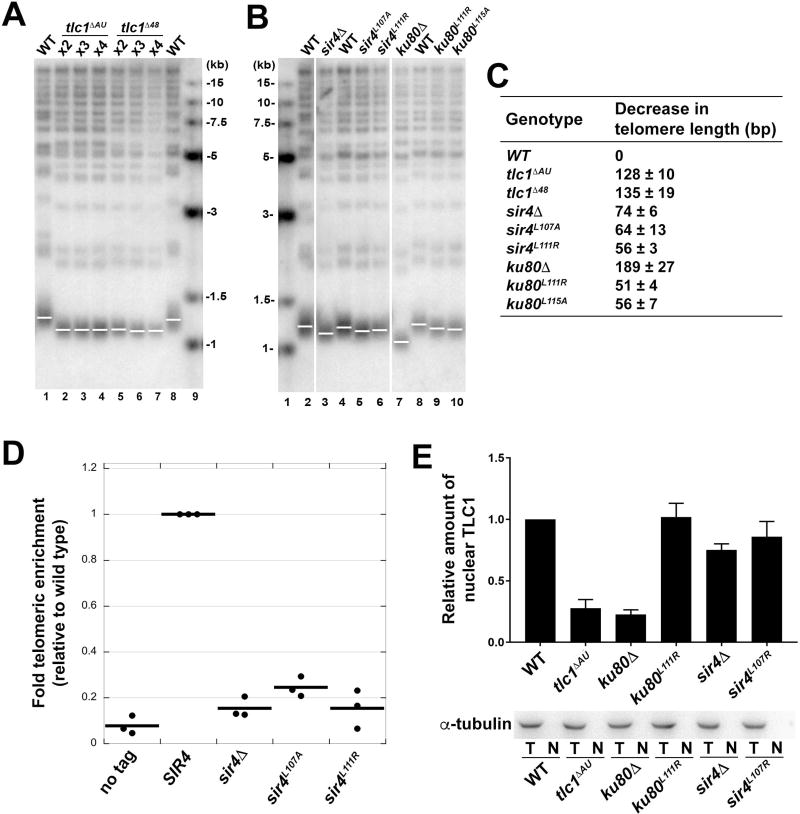
Figure 4. Mutations Disrupting the Ku-TLC1 or Sir4-Ku80 Interactions Reduce Telomere Length.
(A) Southern blot analysis of XhoI-digested yeast genomic DNA shows the length of Y’ telomeres and non-Y’ telomeres over the course of streaks 2–4 using TG(1–3) repeat telomeric probe. (B) Southern blot analysis of telomere lengths of wild-type, sir4 and ku80 mutant strains. Vertical white spaces between lanes 2 and 3, 6 and 7, indicate cropping of superfluous lanes. (C) Measurement of average telomere length of yeast strains shown in (A) and (B). Telomere length was calculated from three independent experiments (mean ± SEM). (D) ChIP analysis of Est2 enrichment at telomere VI-R in wild-type, sir4ΔA, sir4L107A , and sir4L111R strains. The thick horizontal lines on the graphs represent averages of three independent biological replicates indicated by black dots. (E) Upper panel: the quantification of relative TLC1 RNA in nuclei of wild-type, tlclΔAU, ku80A, ku80L111R, sir4Δ, sir4L107R strains. Results are from three independent experiments (mean ± SEM). Lower panel: Western blot of α-tubulin in total cell extracts (T) and nuclear extracts (N) from strains described above indicated the absence of cytoplasmic contaminants in the isolated nuclei.
See also Figure S4.