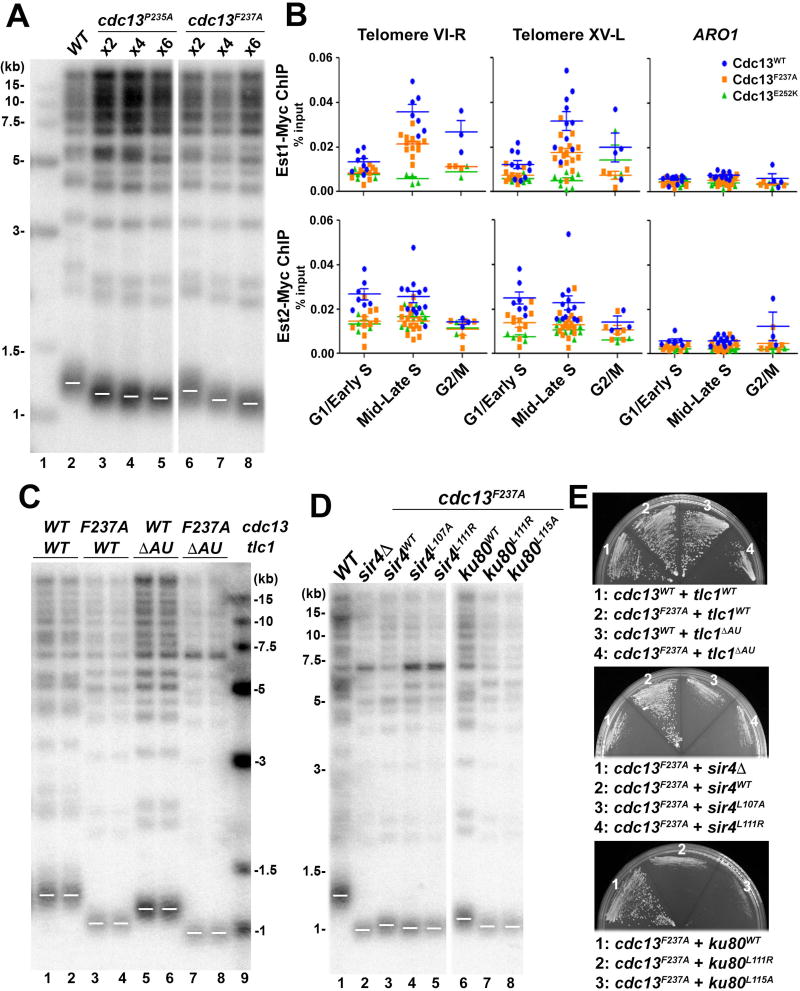
Figure 6. The TLC1-Ku70/80-Sir4 and the Est1-Cdc13 Pathways Have Additive Effect on Telomere Length Regulation.
(A) Mutations in Cdc13EBM-N (cdc13P235A and cdc13F237A) confer reduced but stable telomere length. Cells were taken from serial streaks 2, 4 and 6 after sporulation. Vertical white space between lanes 5 and 6 indicates cropping of superfluous lanes. (B) ChIP assay of Est1 and Est2 enrichment at telomeres VI-R and XV-L in wild-type, cdc13F237A, cdc13E252K strains. Recruitment to the internal ARO1 locus serves as a control of ChIP specificity. Each data point represents the outcome of individual ChIP reaction for at least three cell cycle synchronization experiments (biological replicates). The horizontal lines represent the means and the error bars are standard errors of the means. (C) Strains with both cdc13F237A and tlc1ΔAU mutations (lanes 7–8) exhibited further telomere shortening than single mutation of either cdc13F237A(lanes 3–4) or tlc1ΔAU (lanes 5–6). (D) Strains with both cdc13F237A and mutations that disrupt the Ku80-Sir4 interaction exhibited further telomere shortening. Vertical white space between lanes 5 and 6 indicates cropping of superfluous lanes. (E) Strains as described in (A), (C) and (D) were streaked onto plates to analyze senescence phenotype.
See also Figure S7.