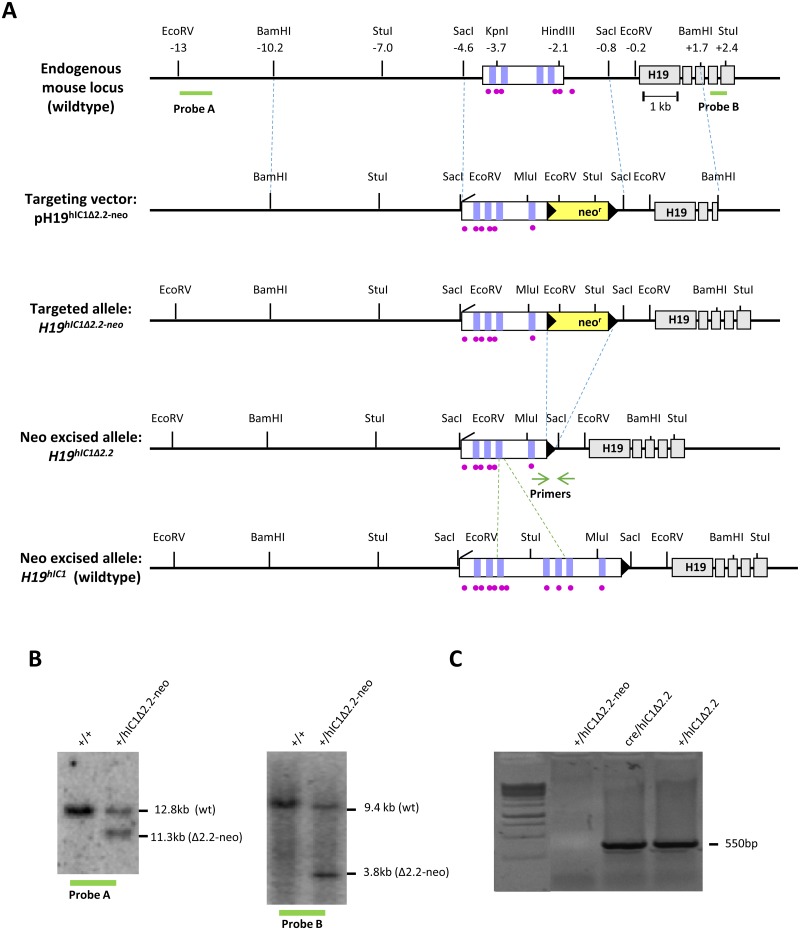
Fig 1. Strategy for generating the H19hIC1Δ2.2 allele.
(A)Schematics of the endogenous mouse H19 locus, targeting vector for mutant hIC1 (pH19hIC1Δ2.2-neo), targeted mutant allele with the neomycin resistance cassette (H19hIC1Δ2.2-neo), targeted mutant allele after neoR excision (H19hIC1Δ2.2) and targeted wildtype hIC1 allele (H19hIC1). Depicted are the IC1 (white rectangle) with CTCF binding sites (vertical lilac bars within IC1), ZFP57 binding sites (purple circles), H19 exons (gray rectangles); pBluescriptIIKS vector with the human insert flanked by the mouse sequences (outlined with dashed blue lines) used as arms for the homologous recombination, neoR cassette (yellow rectangle), loxP sites (black arrowheads). Restriction sites and their relative positions (in kb) to the 5’ end of H19 are also indicated. Probes (A and B) used for Southern blot analysis are indicated as thick green lines below the endogenous locus. Positions of the primers for testing the neo cassette excision are indicated by green arrows. Dashed green lines define the region of hIC1 deleted in the hIC1Δ2.2 allele. (B) Southern blot analysis to confirm targeting of mutant hIC1 allele. DNA from wildtype (+/+) and targeted (+/hIC1Δ2.2-neo) ES cells was digested with EcoRV and hybridized to probe A (lying 5’ of the targeting arm) or digested with StuI and hybridized to probe B (lying 3’ of the targeting arm). (C) Confirmation of Cre-mediated excision of neoR cassette. Excision was confirmed by PCR with primers (green arrows in fig 1A) flanking neoR. A PCR product of 550 bp confirming excision was obtained from genomic DNA of F1 (Cre/hIC1Δ2.2) mice, and F2 (+/hIC1Δ2.2) derived from F1 crossed to wildtype mice. No PCR product was obtained from KI mice (+/hIC1Δ2.2-neo) with the NeoR cassette.