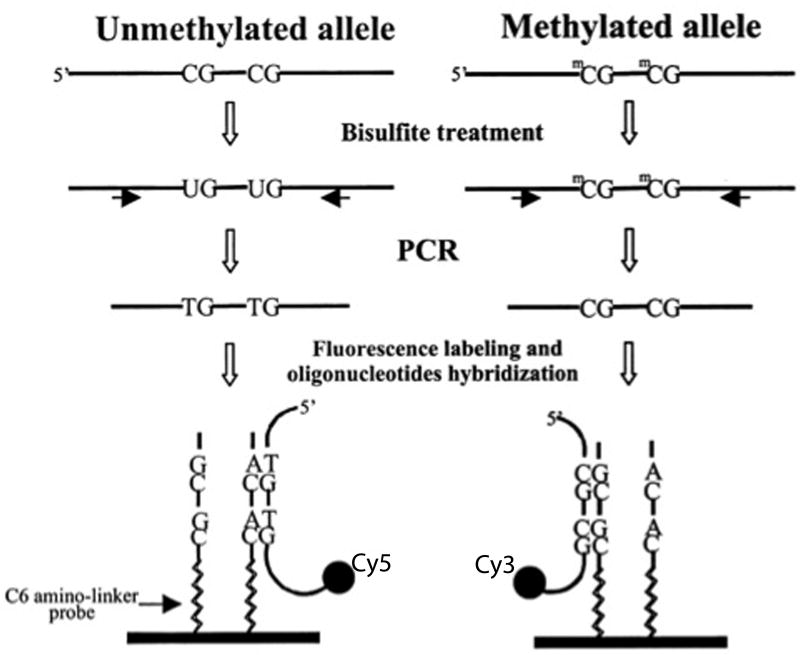
Figure 3.
Following amplification of the region of interest by polymerase chain reaction (PCR), array-based methods rely on hybridization of a sample of deoxyribonucleic acid (DNA) to oligonucleotides contained within the array. Sodium bisulfite treatment of DNA produces a change in the nucleotide sequence of all unmethylated cytosines. Amplification of a specific locus of interest is performed, and a “tag” is incorporated into the PCR products using modified PCR primers that feature one of two unique sequence tails (i.e., one primer detects the methylated state and has one specific tag, while the other primer detects the unmethylated state and has a different tag). This procedure results in fluorescently labeled PCR products with a nucleotide sequence specific to its methylation status. A set of two oligonucleotides is synthetically made with their sequences complementary to the unmethylated and methylated sequence of the labeled PCR products and is attached to the surface of a glass slide. These immobilized oligonucleotides serve as a target for the labeled PCR products and are referred to as probes. The labeled PCR products are incubated with an array of immobilized probes to permit complementary sequences to hybridize to a given probe. A high-resolution camera captures the position of emitted fluorescence. The difference in signal intensities between the paired methylated and unmethylated alleles is used to calculate the percentage of methylation for the sequence associated with the probe. Modified from “Methylation-specific oligonucleotide microarray: A new potential for high-throughput methylation analysis,” by R. S. Gitan, H. Shi, C.-M. Chen, P. S. Yan, & T. H.-M. Huang, Genome Research, 12, p. 159. Copyright 2002 by Cold Springs Harbor Laboratory Press.