Figure 1.
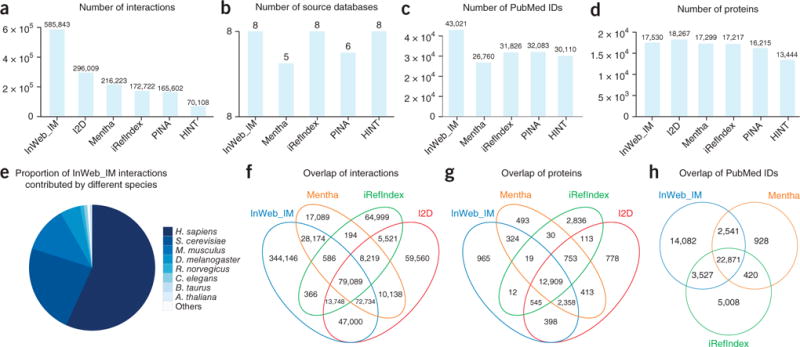
A quantitative comparison of InWeb_IM and five widely used human protein–protein interaction networks. (a–d) Shown are comparisons of the total amount of interactions (a), the number of source databases (b), the number of PubMed identification numbers (c) and the number of proteins (determined using the UniProt reference proteome; d) covered by at least one interaction for each of the six networks. (e) The proportion of InWeb_IM interactions that have been found in humans. (f–h) In the four networks containing the most data, we also measured the overlap of interactions (f), the overlap of proteins covered by at least one interaction (g) and the overlap of publications supporting the interaction data (h) (I2D was excluded in c and h because PubMed identification numbers could not be traced).
