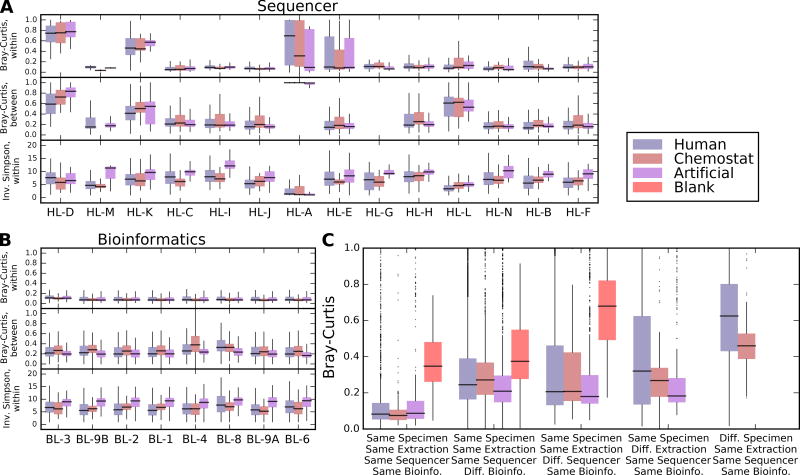
Figure 3. Individual and aggregate effects of sample handling and bioinformatics labs on microbial profiles.
A) Distributions of within- and between-sample alpha and beta diversities, respectively, stratified by sample type (n=2,033 for artificial communities, n=11,991 for human-derived samples, and n=1,725 for chemostat samples) and by handling or B) bioinformatics lab. Raw data, including sample sizes, are included in Supplementary Dataset 7. Bray-Curtis dissimilarities within labs are computed only between technical replicates handled and extracted identically; between lab distributions compare only replicates from the same originating specimen as processed by one lab to all others. Outlier values outside 1.5 times the interquartile range are omitted for clarity. Within-lab comparisons thus assess the consistency of each lab between replicates; between-lab comparisons assess how (dis)similar each lab’s results are to all others. C) Effect size distributions of technical variation (between identically handled replicate samples), differences only due to bioinformatics lab, sequencing lab, extraction (local vs. central), and between different biological specimens. In general, biological differences were largest, followed by extraction (particularly for heterogeneous human-derived samples), sequencing protocol, and computational protocol effects were smallest. Omnibus tests for differences among specimen type, handling laboratory, and bioinformatics laboratory are all significant at Kruskal-Wallis p<0.05; pairwise Wilcoxon tests for the effects of most individual handling laboratories are significant, while most bioinformatics laboratories are not (Supplementary Table 6).