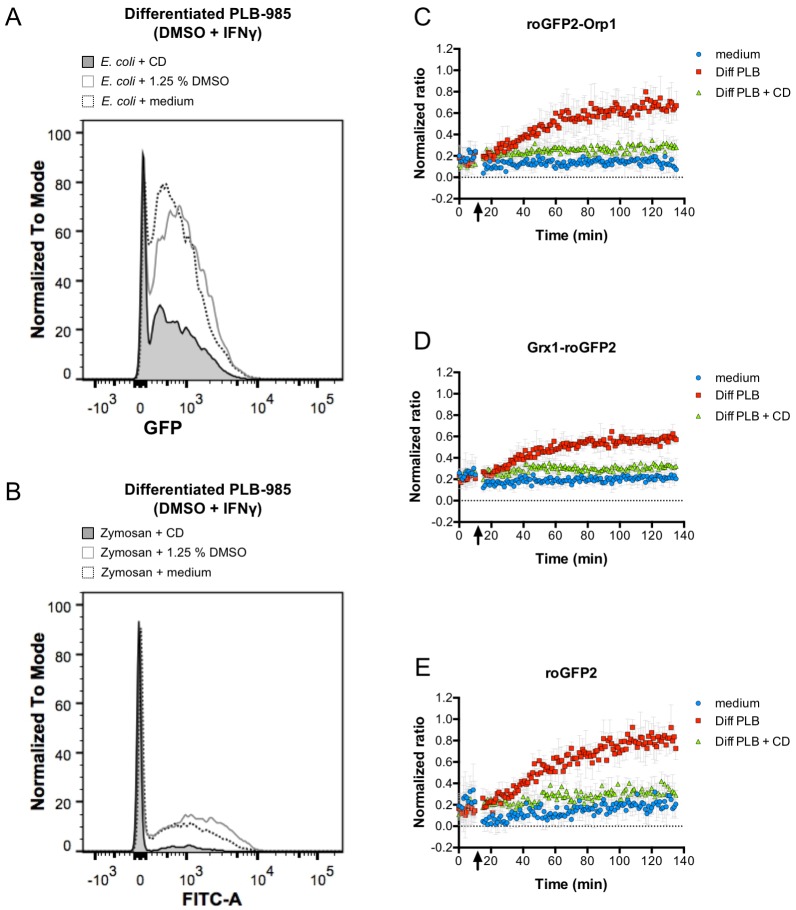
Figure 5. Phagocytosis is required for efficient oxidation of roGFP2-based probes.
Cytochalasin D inhibits phagocytosis of E. coli (A) and FITC-labeled Zymosan (B) as determined by flow cytometry. Cytochalasin D (CD) treatment of differentiated PLB-985 cells inhibits oxidation of roGFP2-Orp1 expressed in E. coli (C). Similar findings were observed in bacteria expressing Grx1-roGFP2 (D) and unfused roGFP2 (E). Arrows indicate the addition of medium, differentiated, neutrophil-like PLB-985 cells and cytochalasin D-treated differentiated PLB-985 cells.