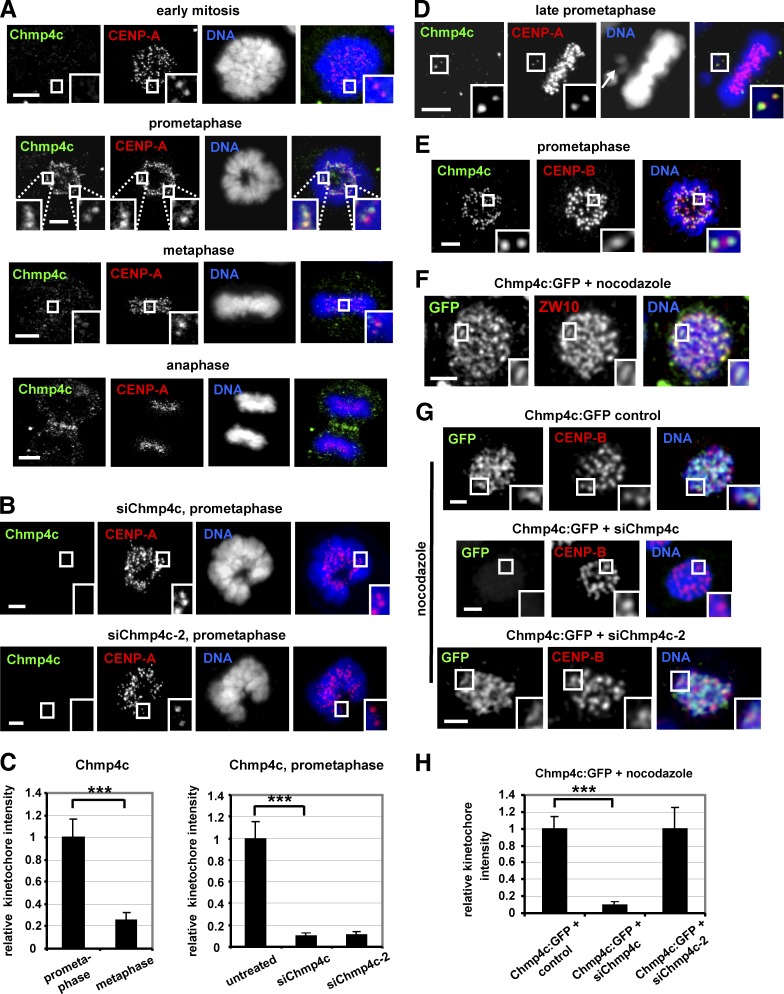
Figure 1.
Chmp4c localizes to kinetochores. (A–C) Cells were untreated or transfected with two different Chmp4c siRNAs (siChmp4c and siChmp4c-2). Relative green/red fluorescence intensity is shown, and values in prometaphase or untreated were set to one. (D) Example of a late-prometaphase cell exhibiting an unaligned chromosome (indicated by an arrow) and an acentric chromatin fragment. Chmp4c localizes to unaligned kinetochores (insets) but is diminished from kinetochores at the metaphase plate. (E) Localization of Chmp4c and CENP-B. (F) Localization of Chmp4c:GFP in cells treated with nocodazole for 4 h. (G and H) Localization of Chmp4c:GFP, resistant to degradation by siChmp4c-2, in cells transfected with negative siRNA (control), siChmp4c, or siChmp4c-2 and treated with nocodazole for 4 h. Relative green/red fluorescence intensity is shown, and values in the control were set to one. n > 200 kinetochores, 20 cells (C and H) from three independent experiments. Error bars show the SD. ***, P < 0.001 compared with the control. Statistical significant differences were determined by ANOVA and Student’s t test. Insets show 1.7× magnification of kinetochores. Bars, 5 µm.