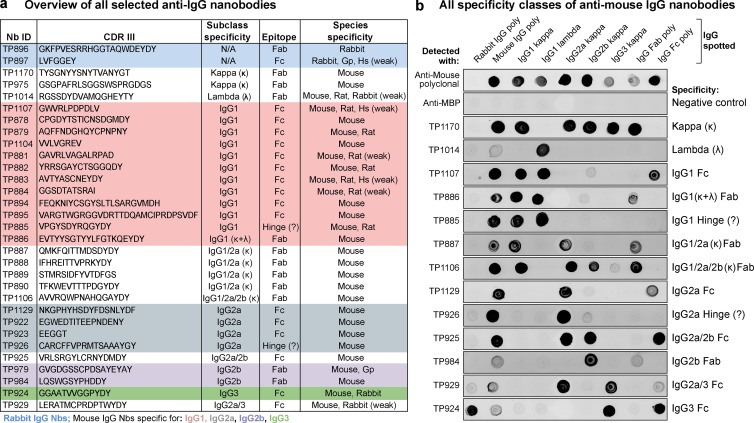
Figure 1.
Characterization of the anti-IgG nanobody toolbox. (a) Overview of all identified anti-IgG nanobodies. The nanobodies obtained were characterized for IgG subclass and light chain specificity, epitope location on Fab or Fc fragment, and species cross reactivity. The protein sequences of all anti-IgG nanobodies can be found in Table S1. Nb, nanobody; CDR III, complementarity-determining region III; Gp, guinea pig; Hs, human; κ, κ light chain; λ, lambda light chain; Fab, fragment antigen-binding, Fc, fragment crystallizable. (b) IgG subclass reactivity profiling of selected anti–mouse IgG nanobodies representing all identified specificity groups. The indicated IgG species were spotted on nitrocellulose strips, and the strips were blocked with 4% (wt/vol) milk in 1× PBS. Then 300 nM of the indicated tagged nanobodies were added in milk. After washing with 1× PBS, bound nanobodies were detected using a fluorescence scanner. Note that the signal strength on polyclonal IgG depends on the relative abundance of the specific subclass (e.g., IgG2b and IgG3 are low abundance) or light chain (κ/λ ratio = 99:1). TP885 and TP926 showed no detectable binding to polyclonal Fab or Fc fragment and might bind to the hinge region. MBP, maltose binding protein; poly, polyclonal.