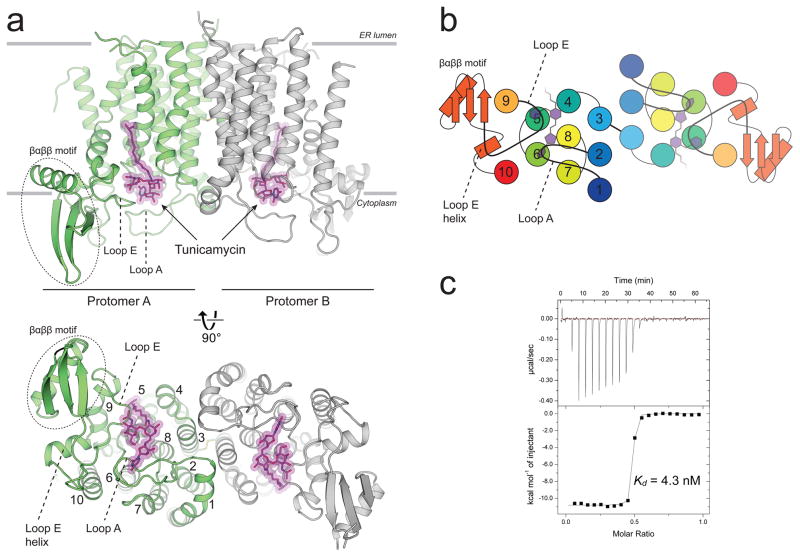
Figure 1. The crystal structure of human GPT in complex with tunicamycin reveals the structural basis for high affinity binding.
a, hGPT crystallized as a homodimer with one tunicamycin molecule (magenta sticks) bound to the cytosolic active site cavity of each protomer, tightly enclosed by loops A and E. The two protomers are related by pseudo-twofold rotational symmetry and are covalently linked by a disulfide bond between TM3 of each protomer. b, Cartoon representation of the cytoplasmic view, rainbow colored from N-terminus (blue) to C-terminus (red). c, Representative ITC raw data (top) and binding isotherm (bottom) for tunicamycin interacting with wild-type hGPT; Kd = 4.3 nM, ΔH° = −10.8 kcal/mol. This ITC experiment was performed in triplicate (technical replicates (5.6+/−1.3 nM, mean +/− S.E.M.)).