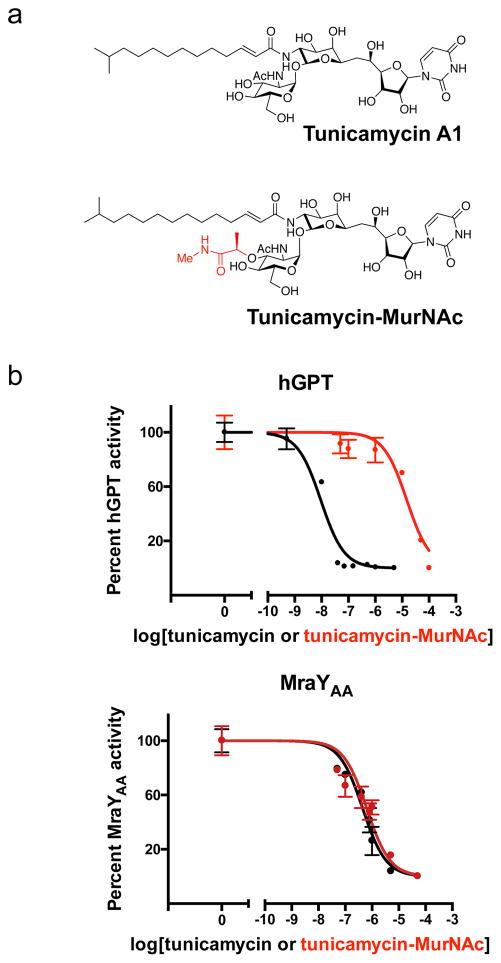
Figure 5. Chemically modifying tunicamycin can introduce ligand selectivity between hGPT and MraYAA.
a, Chemical structures of tunicamycin and its MurNAc derivative. The substructure highlighted in red differs from tunicamycin, being a MurNAc-like moiety rather than a GlcNAc moiety as in tunicamycin. b, IC50 measurements of tunicamycin and its MurNAc analog (tunicamycin-MurNac) with hGPT and MraYAA, respectively. hGPT is much less inhibited by tunicamycin-MurNAc than tunicamycin, while MraY appears to be similarly inhibited by both tunicamycin and tunicamycin-MurNAc. The IC50 for hGPT with tunicamycin is ~9 nM; for hGPT with tunicamycin-MurNAc is ~15 μM; for MraYAA with tunicamycin is ~450 nM; for MraYAA with tunicamycin-MurNAc is ~640 nM. Each IC50 value was determined using the TLC-based phosphoglycosyl transferase assay. Data are shown as the mean of triplicate measurements (technical replicates) ± s.e.m.