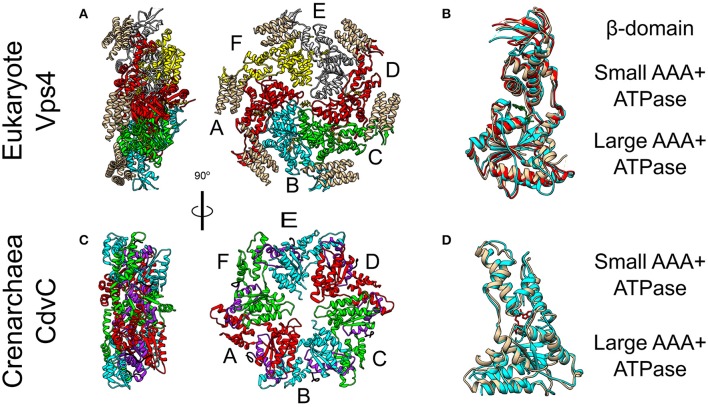
Figure 5.
Crystal Structure of the Vps4 and CdvC proteins. (A) Crystal structure in side (left) and front (right) views of the yeast Vps4 hexamer together with the VSL domain of Vta1 (tan). Protomers are marked as A–F. Equivalent monomers share color. Gray protomers bind either ADP or ADP+Pi. The yellow protomer is empty (PDB #5UIE) (Monroe et al., 2017). (B) Alignment of monomers F (tan), E (red) and C (cyan). F to C protomers alignment—RMSD between 300 atom pairs is 3.262 Å. (C) Crystal structure side (left) and front (right) views of an empty M. sedula CdvCΔMIT hexamer (PDB #4D80) (Caillat et al., 2015). Identical protomers share the same color. The P-Loops NTPase domains (residues 105–156) are highlighted in purple. The N-terminus of every chain is shown in black. (D) Alignment of one protomer from (C) in tan with M. sedula CdvC bound to ADP (PDB #4D82) in cyan. RMSD between 257 atom pairs is 2.717 Å.