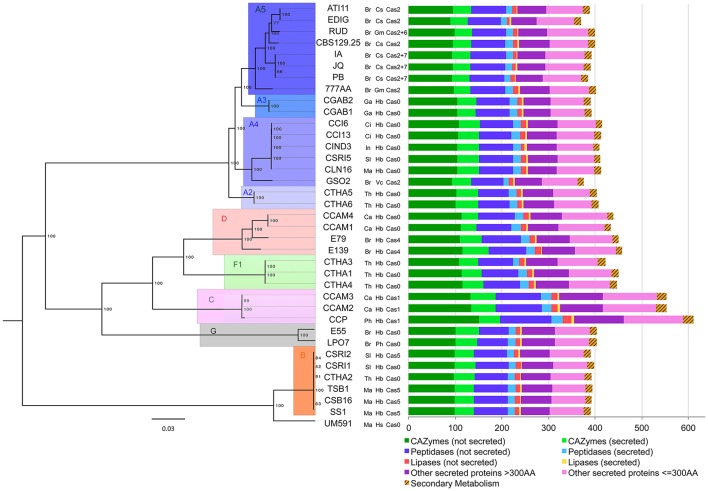
Figure 5.
Intraspecific phylogenomic tree and putative accessory effectors of C. cassiicola isolates. The maximum likelihood phylogenomic tree was based on 12,420 conserved protein sequences. Branch lengths are indicated by the bar (substitutions/site); 1,000 bootstrap values are shown as percentages. The isolates names are followed by their country and host codes, and by the toxin class, as indicated in Table 1. The diagram on the right represents the composition in putative accessory effectors (i.e. either absent or varying in copy number in at least one isolate).