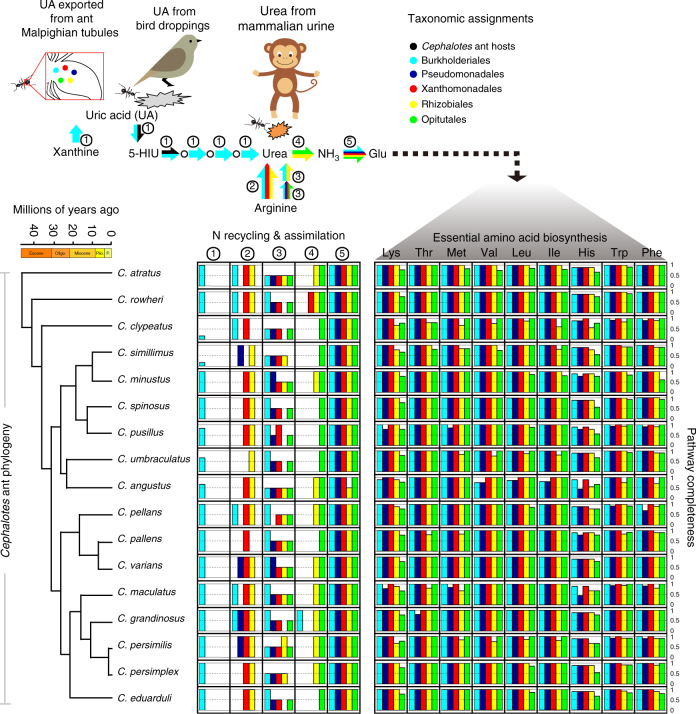
Fig. 4.
Pathways for N-waste recycling and amino acid biosynthesis and their distributions across core gut symbionts from 17 Cephalotes species. Various symbiotic gut bacteria convert N wastes into ammonia, incorporate ammonia into glutamate, and synthesize essential amino acids. The proposed model in the upper panel shows sources of the N-waste uric acid (bacterial metabolism via xanthine degradation, bird droppings, host ant waste metabolism via Malpighian tubule delivery) and urea (mammalian urine, uric acid metabolism, and arginine metabolism). Arrows in this panel are colored to reflect taxonomy of the core Cephalotes-specific microbes participating in these steps in multiple metagenomes. Numbers near arrows link particular pathways to bar graphs (below), which in turn plot pathway completeness (i.e., proportion of all genes present) for the dominant core taxa in each metagenome. At left on the lower panel is the phylogeny of Cephalotes species used for metagenomics including a chronogram dating divergence events in these species’ history27. The bird and monkey images used in the upper panel were created using Adobe Illustrator CS6 (v. 16.0.0) and the ant image is an Adobe Illustrator clipart image