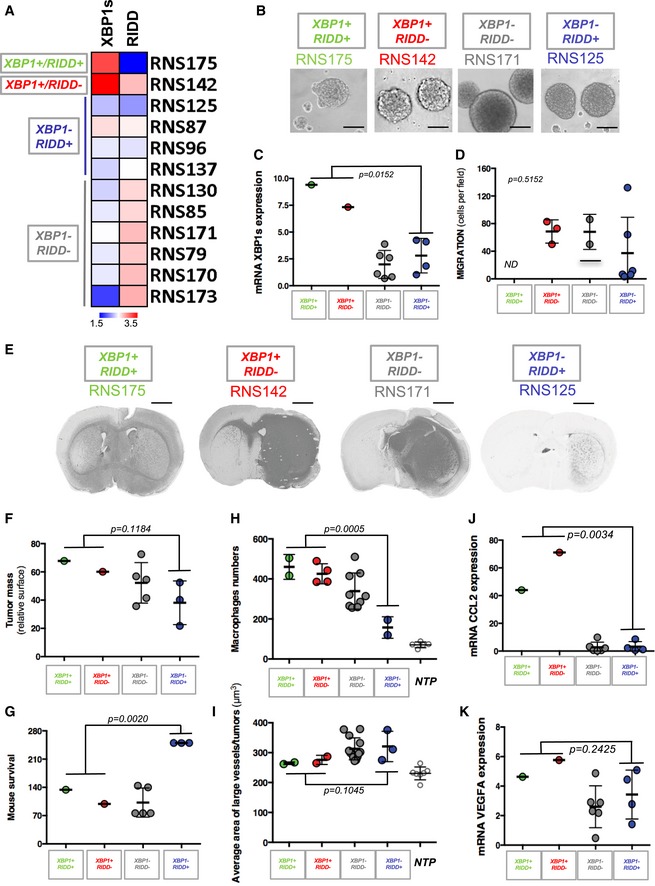
-
A
Hierarchical clustering of 12 GBM primary lines based on XBP1s and RIDD scores (see
Materials and Methods).
-
B
Phase contrast images of the phenotypes exhibited by the primary lines from the four groups established in (A) (scale bar = 100 μm).
-
C
Quantitation of XBP1s mRNA in all primary lines relative to the group to which they belong (n = 3, mean ± SD). ANOVA was used for statistical analyses.
-
D
Quantitation of the Boyden chamber migration assays for all the lines (n = 3, mean ± SD). ANOVA was used for statistical analyses.
-
E
GBM primary cell lines were implanted in nude mice. Animals were sacrificed when first clinical signs appeared. Brains were then collected and analyzed for vimentin expression by immunohistochemistry (scale bar = 800 μm).
-
F
Quantitation of tumor size. ANOVA was used for statistical analyses. For each cell line tested (represented by a single point on the graph), the average of three independent experiments is shown ±SD.
-
G
Quantitation of mouse survival. ANOVA was used for statistical analyses. For each cell line tested (represented by a single point on the graph), the average of three independent experiments is shown ±SD.
-
H
Quantitation of macrophage infiltration in orthotopic tumors (IBA1 staining). ANOVA was used for statistical analyses. For each cell line tested (represented by a single point on the graph), the average of three independent experiments is shown ±SD.
-
I
Quantitation of angiogenesis in orthotopic tumors (CD31 staining). ANOVA was used for statistical analyses. For each cell line tested (represented by a single point on the graph), the average of three independent experiments is shown ±SD.
-
J, K
Expression of CCL2 (J) and VEGF‐A (K) mRNA as determined using RT‐qPCR in the different primary GBM lines. ANOVA was used for statistical analyses. For each cell line tested (represented by a single point on the graph), the average of three independent experiments is shown ±SD.