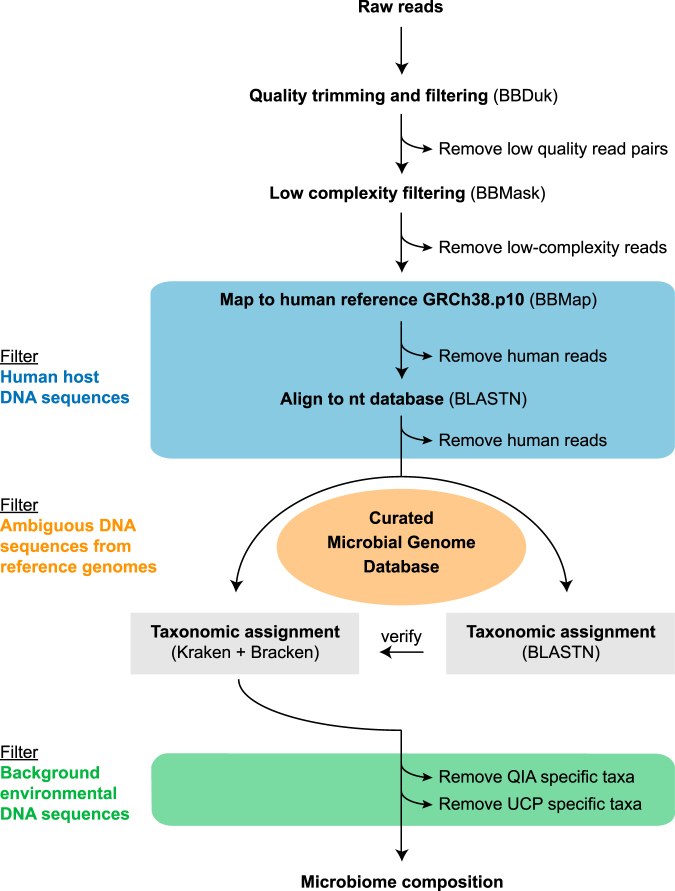
Figure 2.
Workflow for metagenomic data analysis. In a first step, sequencing adapters, low quality bases and reads with low complexity were removed. Subsequently, reads that mapped against the human reference genome sequence, or aligned with human sequences in the nt database were removed. The taxonomic classification of the reads was performed with Kraken together with Bracken using a curated microbial genome database containing 5,750 microbial (archaea [251], bacteria [5,166], fungi [225], protozoa [73], viruses [35]) and 1 human reference genome sequence (for details, see Supplementary Methods). Additional reads that in this step were classified as human were removed. To verify the classification results, the reads were also aligned to the reference genomes using BLASTn. Organisms specific for the DNA extraction (blank) controls were filtered from the patient samples.