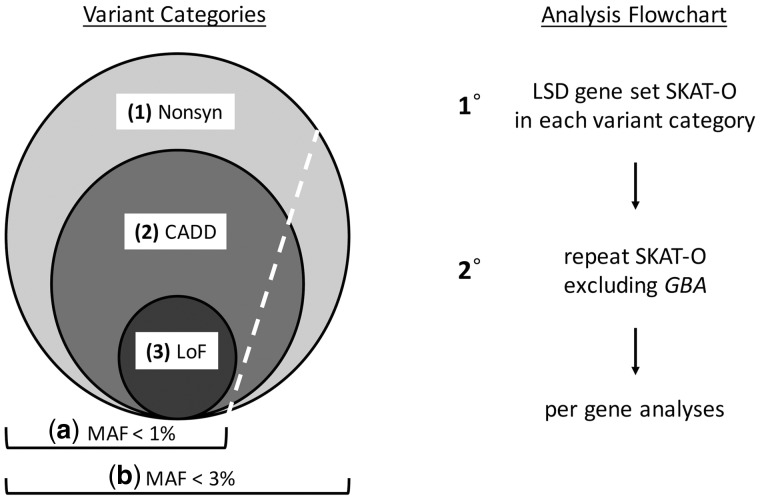
Figure 1.
Overall analytic strategy. (Left) Variant categories. Because the number, frequency, and effect sizes of Parkinson’s disease risk variants remains incompletely defined, our analyses considered three nested categories based on increasing variant pathogenicity: (1) all non-synonymous variants (Nonsyn); (2) likely damaging variants based on combined annotation dependent depletion (CADD) score; and (3) loss-of-function (LoF) variants. Based on the known prevalence of Parkinson’s disease and incomplete penetrance documented for many risk alleles, we also considered two frequency thresholds, including rare (MAF < 1%) and somewhat more common (MAF < 3%) variants. (Right) Analysis flow chart. The SKAT-O was initially performed for the complete LSD gene set in the IPDGC discovery cohort, considering each variant category separately. For those categories with a significant SKAT-O association in the full gene set, a secondary analysis was performed excluding all GBA variants in order to confirm the involvement of additional genes. This was repeated in each of the replication cohorts (PPMI and NeuroX). Lastly, to highlight those loci driving associations detected in the gene set, secondary analyses were performed in the IPDGC cohort using SKAT-O to evaluate variants in each LSD gene independently.