Fig. 8.
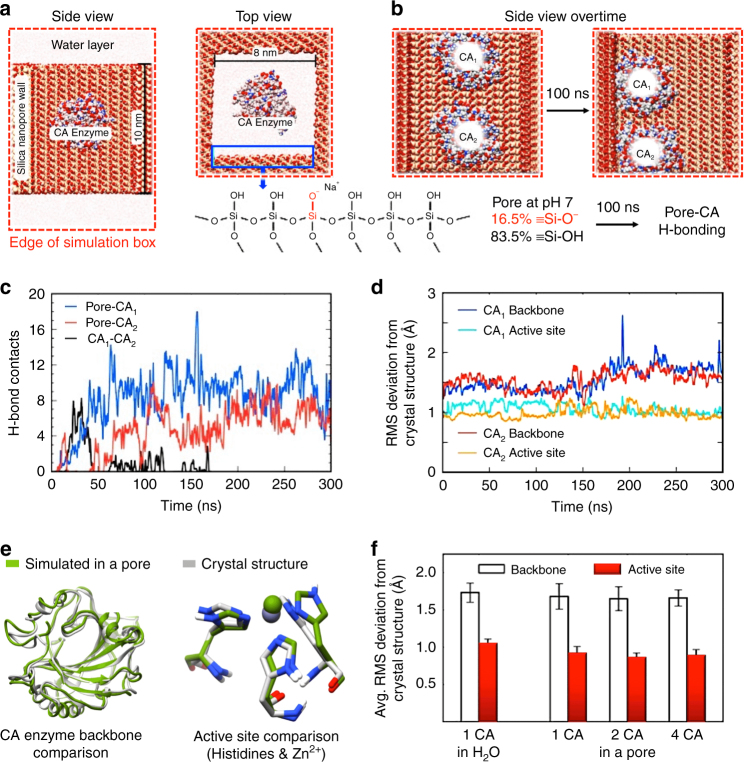
Molecular dynamics simulations of carbonic anhydrase confinement within individual mesopore channels of the enzymatic liquid membrane. a, b Setup for the molecular dynamics simulations of CA enzymes confined within a silica nanopore. c CA enzymes rapidly (<100 ns) adsorb to the surface of the silica nanopore and remain in contact with the pore walls for the duration of the simulation, as shown by H-bond contacts between the pore and CA in c. d Root-mean-squared deviation (RMSD) data of the protein backbone and active site atoms shows that the structure of the CA enzymes adsorbed to the pore remains stable during the course of the simulation and close to the crystal structure. e Ribbon representation (left) and close-up of the active site of the CA enzyme show close similarity between the average structure simulated in the nanopore and crystal structure. f Average RMSD data (over the last 50 ns of 300 ns simulations) for 1, 2, and 4 CA enzymes per pore (to simulate varying crowded conditions) shows that the structure of the enzyme is highly robust and resembles that of the free CA structure in solution. Error bars represent 95% confidence intervals for experiments performed with n = 3