Insertional mutagenesis of mannosidase 1A and xylosyltransferase 1A results in altered N-glycan compositions, indicating diverse roles in the trimming and modification of N-glycans in C. reinhardtii.
Abstract
At present, only little is known about the enzymatic machinery required for N-glycosylation in Chlamydomonas reinhardtii, leading to the formation of N-glycans harboring Xyl and methylated Man. This machinery possesses new enzymatic features, as C. reinhardtii N-glycans are independent of β1,2-N-acetylglucosaminyltransferase I. Here we have performed comparative N-glycoproteomic analyses of insertional mutants of mannosidase 1A (IMMan1A) and xylosyltransferase 1A (IMXylT1A). The disruption of man1A affected methylation of Man and the addition of terminal Xyl. The absence of XylT1A led to shorter N-glycans compared to the wild type. The use of a IMMan1AxIMXylT1A double mutant revealed that the absence of Man1A suppressed the IMXylT1A phenotype, indicating that the increased N-glycan trimming is regulated by core β1,2-Xyl and is dependent on Man1A activity. These data point toward an enzymatic cascade in the N-glycosylation pathway of C. reinhardtii with interlinked roles of Man1A and XylT1A. The results described herein represent the first step toward a functional characterization of the enzymatic N-glycosylation machinery in C. reinhardtii.
The diverse roles of N-glycosylation, a major and essential posttranslational protein modification, are closely linked to the respective N-glycan structures as well as to the protein the modification is attached to. While the initial steps of the N-glycosylation pathway in the endoplasmic reticulum (ER), such as the synthesis and transfer of the lipid-linked oligosaccharide Glc3Man9GlcNAc2 to a nascent polypeptide, are conserved in most eukaryotes, trimming and maturation of the N-glycan in the Golgi apparatus can be highly variable. After removal of all Glc residues and one Man residue in the ER, the N-glycan is further trimmed by class I α-mannosidases in the Golgi apparatus. In plants, the addition of β1,2-GlcNAc (GlcNAc) by β1,2-N-acetylglucosaminyltransferase I (GnTI) is required for the formation of complex N-glycans that can be further modified by α-mannosidase II and GnTII. Typically, complex plant N-glycans harbor a core β1,2-Xyl and/or α1,3-Fuc and can be terminally capped by β1,3-Gal and α1,4-Fuc residues, resulting in Lewisa epitopes. The biological importance and diverse functions of N-glycosylation have been reviewed and can be illustrated by various phenotypes of knockout mutants in the N-glycosylation pathway (Strasser 2016). In Arabidopsis (Arabidopsis thaliana), mutants lacking Lewisa structures were not affected in their growth or development (Strasser et al., 2007, 2008). In contrast, strains lacking core Fuc and/or Xyl were hypersensitive toward salt stress (Kang et al., 2008). While the knockout of GnTI in Arabidopsis merely resulted in reduced growth under salt stress (Kang et al., 2008), a lack of GnTI in rice (Oryza sativa) severely affected its growth under normal conditions and led to early lethality (Fanata et al., 2013). Regarding class I α-mannosidases in Arabidopsis (MNS1-3), MNS3 catalyzes the removal of one Man from Man9GlcNAc2 and MNS1/2 cleave off three Man from Man8GlcNAc2. Single knockout mutants did not exhibit apparent phenotypes, but a triple mns mutant showed a root growth phenotype and altered cell wall (Liebminger et al., 2009).
The N-glycosylation pathway of Chlamydomonas reinhardtii has been revealed only recently (Mathieu-Rivet et al., 2013, Vanier et al., 2017). Thereby, novel types of N-glycans were identified as well as uncommon linear lipid-linked oligosaccharides. Additionally, and in contrast to vascular plants, no GnTI homolog was found in the genome of C. reinhardtii and N-glycans were lacking GlcNAc on both branches. Nevertheless, they still harbored β1,2-Xyl and Fuc at the N-glycan core. Additionally, a second, terminal Xyl could be attached and N-glycans were modified with 6-O-methylated Man residues, which had been reported previously for Porphyridium sp. (Levy-Ontman et al., 2011). In silico analysis of the C. reinhardtii genome revealed candidates for all major enzymes of the N-glycosylation pathway (Mathieu-Rivet et al., 2013). However, the activity and specificity of these candidates has not been shown so far.
C. reinhardtii is a model organism, for example for flagellar biogenesis, photosynthesis, and acclimation to nutrient deficiency (Harris et al., 2009; Merchant et al., 2007). Understanding the role and biosynthetic pathway of its N-glycosylation is important for the analysis of these processes. This is emphasized by the identification of N-glycoproteins involved in for example iron assimilation, iron assimilating protein 1/2 and ferroxidase 1, or homologs to human proteins associated with polycystic kidney disease, polycystic kidney disease 2 (Mathieu-Rivet et al., 2013).
Here, we performed a comparative N-glycoproteomic analysis of various N-glycosylation pathway mutants in C. reinhardtii. This study includes the identification and characterization of insertional mutants for (1) α-mannosidase 1A (IMMan1A), which has been proposed to function as the only class I α-mannosidase in the trimming of N-glycans in the ER of C. reinhardtii, and (2) xylosyltransferase 1A (IMXylT1A), which has been predicted to be the only β1,2-xylosyltransferase (XylT) in C. reinhardtii (Mathieu-Rivet et al., 2013), as well as (3) a double mutant that was generated by genetic crossing of the two insertional mutant strains. Each of these mutant strains exhibited a distinct, altered N-glycan composition, indicating different roles for Man1A and XylT1A in the regulation of N-glycan trimming and maturation.
RESULTS
Insertional Mutant Library Screening and Comparative N-Glycoproteomics
To modify N-glycan structures in C. reinhardtii, an insertional mutant library was screened for mutants in the N-glycosylation pathway and insertions in genes encoding for Man1A and XylT1A were identified (Cheng et al., 2017). These mutants were genetically crossed and glycopeptides from single and double mutants were analyzed by mass spectrometry (MS).
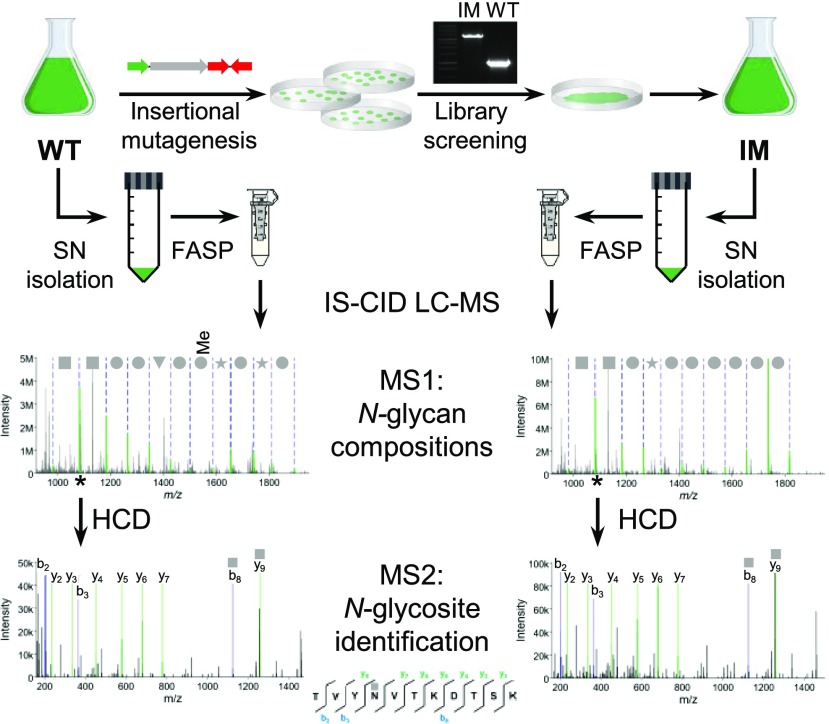
For the analysis of intact, nonenriched N-glycopeptides, in-source collision-induced dissociation (IS-CID) was employed as described previously (Mathieu-Rivet et al., 2013), which mainly leads to the fragmentation of glycosidic linkages on MS1-level. Thus, N-glycan compositions can be deduced by a series of neutral losses. Using mass tags for the selection of ions differing by the mass of one N-acetylhexosamine (HexNAc), peptides carrying one or two HexNAc can be designated for further fragmentation via higher-energy collisional dissociation to deduce the peptide sequence corresponding to the respective N-glycan. With this approach, N-glycan compositions for the same N-glycosites in different strains can be compared (Fig. 1).
Figure 1.
Schematic representation of the workflow used for the comparative N-glycoproteomic analysis of the wild type (WT) and insertional mutant strains. After identification of IM strains for enzymes involved in the N-glycosylation pathway of C. reinhardtii, proteins from the culture SN of the wild type were digested using the FASP method. Subsequently, intact N-glycopeptides were analyzed by mass spectrometry employing IS-CID, allowing for the analysis of the N-glycan composition on MS1 level, while further fragmentation by higher-energy collisional dissociation led to the identification of the peptide sequence on MS2 level.
In general, N-glycan compositions of the wild type and backcrossed strains lacking the insert (see below) were in accordance with what has been described previously (Mathieu-Rivet et al., 2013) including methylated hexose (MeHex) residues, deoxyhexose (dHex) as well as core and terminal pentose (Pent). While these results illustrate the validity of the approach, it should be noted that they do not yield information about the relative abundance of different N-glycans. Nevertheless, since intact N-glycopeptides have been analyzed, the number of N-glycosites harboring specific N-glycan compositions can be determined for each strain. However, it should be noted that the abundance of N-glycans released by peptide-N-glycosidase A/F, as described previously (Mathieu-Rivet et al., 2013, Vanier et al., 2017), is not correlated to the number of different peptides bearing these glycans. In addition, these former analyses were performed on total cell extracts, while secreted proteins have been analyzed here. This might also explain the high degree of N-glycopeptide modifications, including dHex, which we identified here.
Insertional Mutagenesis of man1A Results in Widely Decreased N-Glycan Modification by MeHex and Terminal Pent
The mutant library screen resulted in the identification of IMMan1A carrying an insert in the man1A gene (Cre07.g336600, previously manI). Integration of the insertional cassette into the seventh exon of man1A was revealed by PCR using a gene-specific primer pair (Supplemental Fig. S1, A and B) and sequencing of the PCR product (Supplemental Data S1). PCR across the insertion site verified that no additional deletions occurred adjacent to the insertion site (Supplemental Fig. S1B). To confirm a disturbed expression of man1A, real time PCR as well as parallel reaction monitoring (PRM) measurements were performed. The results revealed a strong reduction of man1A mRNA beneath the detection limit, which was also confirmed on protein level (Supplemental Fig. S1, C and D).
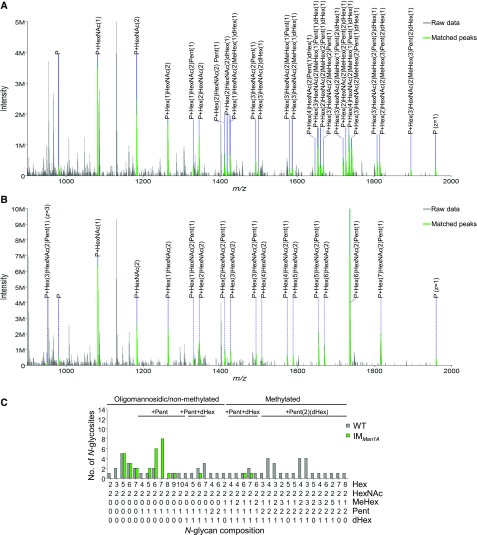
Interestingly, when comparing N-glycans attached to N-glycosites common to IMMan1A and wild type, N-glycans that were methylated and harbored two Pent in the wild type were almost completely devoid of MeHex and only rarely exhibited terminal Pent residues in IMMan1A (Fig. 2). This, to the largest extent, lack of methylation and second Pent was also observed when taking into account all identified N-glycosites, that is not filtering for their occurrence in both strains (Supplemental Fig. S2A; Supplemental Data S2). To confirm the interdependency of the disrupted man1A gene and the altered N-glycan composition, IMMan1A was crossed with wild-type strain CC-124 to obtain the insertional cassette in a different genetic background. Progenies of the crossing were verified by the occurrence of the insert in the man1A gene in a strain encoding for the opposite mating type compared to IMMan1A (CC-124xIMMan1A Ins+; Supplemental Fig. S2C). In line with the parental IM strain, N-glycans of CC-124xIMMan1A Ins+ (mt−) showed less MeHex residues and a clear decrease in the modification by two Pent residues (Supplemental Fig. S2B).
Figure 2.
Comparative analysis of intact N-glycopeptides from wild type and IMMan1A. Applying IS-CID for the analysis of intact N-glycopeptides revealed a decreased number of MeHex and Pent in IMMan1A in comparison to the wild type. Representative MS1 spectra for ITYATTAAAVTNANLSSYK are shown for wild type (A) and IMMan1A (B), respectively. If not indicated otherwise, ions of charge state two are annotated. For all identified N-glycan compositions, the number of N-glycosites harboring this glycan is shown for the wild type (gray) and IMMan1A (green; C). The N-glycan complexity is increasing from left (oligomannosidic, not methylated) to right (decorated, methylated) and N-glycan compositions have been grouped according to the presence of Pent and/or dHex (optional for sugars written in parenthesis). Only N-glycosites that have been identified in both strains were taken into account (n = 26). A comparison for all identified N-glycopeptides can be found in Supplemental Figure S2. Peptide sequences and their corresponding N-glycan compositions are listed in Supplemental Data S2.
Insertional Mutagenesis of xylT1A Results in Decreased N-Glycan Length and Lack of Core Pent
With IMXylT1A, carrying the insertional cassette in the xylt1A gene (Cre09.g391282, previously xylt), a second N-glycosylation pathway mutant could be identified in the mutant library screen. Genetic analyses were performed as for IMMan1A and revealed the location of the insertional cassette in the intron region between exon six and seven (Supplemental Fig. S3, A and B). As shown for IMMan1A, a strong reduction of gene expression was confirmed for IMXylT1A by analysis of transcript and protein level (Supplemental Fig. S3, C and D). Notably, this is in accordance with a strong diminishment of target gene expression after insertion of the same cassette in an intron of the gene encoding for calredoxin (Hochmal et al., 2016).
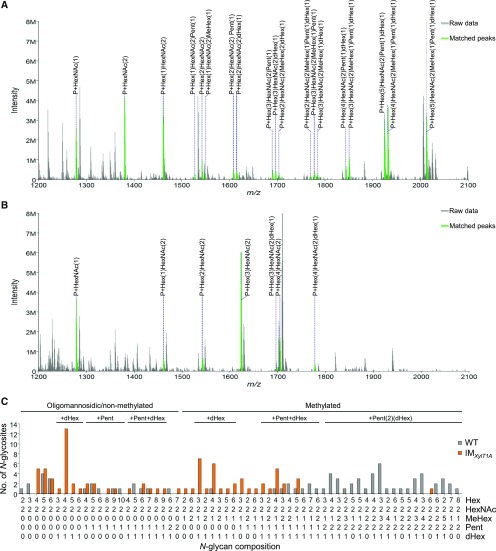
Surprisingly, comparing intact N-glycopeptides of IMXylT1A and wild type revealed a striking decrease in N-glycan length for IMXylT1A (Fig. 3). Indeed, the most common N-glycoform in IMXylT1A comprised only four Hex, whereas the wild type exhibited N-glycans with mainly five to seven Hex. Furthermore, most N-glycans from IMXylT1A carried only one Pent. The almost complete lack of core Pent was also observed when all identified N-glycosites were considered (Supplemental Fig. S4A). As for IMMan1A, mating experiments were carried out using IMXylT1A and CC-124. Progenies showed the same N-glycan composition patterns as the respective parental strains: CC-124xIMXylT1A Ins+ (mt−) exhibited shorter N-glycans and less Pent whereas CC-124xIMXylT1A Ins− (mt+) mainly showed wild-type-like N-glycans (Supplemental Fig. S4, B and C).
Figure 3.
Comparative analysis of intact N-glycopeptides from wild type and IMXylt1A. An increased N-glycan trimming and decreased number of Pent in comparison to the wild type could be observed for IMXylT1A using IS-CID. Representative MS1 spectra for ENSNTTEDGNLFGAAPNVFISR are shown for the wild type (A) and IMXylT1A (B), respectively. Annotated ions have a charge state of two. For all identified N-glycan compositions, the number of N-glycosites harboring this glycan is shown for the wild type (gray) and IMXylt1A (orange; C). The N-glycan complexity is increasing from left (oligomannosidic, not methylated) to right (decorated, methylated) and N-glycan compositions have been grouped according to the presence of Pent and/or dHex (optional for sugars written in parenthesis). Only N-glycosites that have been identified in both strains were compared (n = 33). A comparison for all identified N-glycopeptides can be found in Supplemental Figure S4. Peptide sequences and their corresponding N-glycan compositions are listed in Supplemental Data S2.
Genetical Crossing Leads to an IMMan1AxIMXylT1A Double Mutant Devoid of MeHex and Terminal Pent
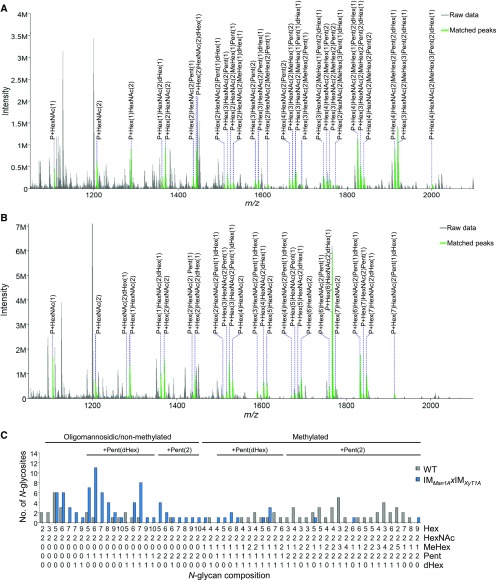
To obtain a Man1A/XylT1A double mutant, the two strains were subjected to mating. After confirming the presence of insertional cassettes in both loci of interest by PCR with the respective insert- and gene-specific primers (Supplemental Fig. S5, C and D), N-glycan compositions of one progeny [IMMan1AxIMXylT1A (mt+)] were analyzed. In line with the lack of Man1A, IMMan1AxIMXylT1A exhibited N-glycan compositions comparable to IMMan1A with a drastic decrease in methylation and the presence of mainly only one Pent residue when comparing N-glycosites common to the wild type and IMMan1AxIMXylT1A (Fig. 4). However, intriguingly, short N-glycans comprised of four Hex, as observed in the majority of N-glycoforms for IMXylT1A, were not detected in IMMan1AxIMXylT1A. Furthermore, the modification of several IMMan1AxIMXylT1A N-glycans with core Pent or even two Pent contrasts with the single mutants as well. These results were also obtained when taking into account all identified N-glycosites (Supplemental Fig. S5A).
Figure 4.
Comparative analysis of intact N-glycopeptides from wild type and IMMan1AxIMXylt1A. Applying IS-CID for intact N-glycopeptide analysis, a decreased number of MeHex and Pent could be observed in IMMan1AxIMXylt1A in comparison to the wild type. Representative MS1 spectra for NQTAINSLVDDIQNTYAK are shown for the wild type (A) and IMMan1AxIMXylt1A (B), respectively. Annotated ions have a charge state of two. For all identified N-glycan compositions, the number of N-glycosites harboring this glycan is shown for the wild type (gray) and IMMan1AxIMXylt1A (blue; C). The N-glycan complexity is increasing from left (oligomannosidic, not methylated) to right (decorated, methylated) and N-glycan compositions have been grouped according to the presence of Pent and/or dHex (optional for sugars written in parenthesis). Only N-glycosites that have been identified in both strains were compared (n = 32). A comparison for all identified N-glycopeptides can be found in Supplemental Figure S5. Peptide sequences and their corresponding N-glycan compositions are listed in Supplemental Data S2.
Comparison of N-Glycan Compositions in Wild Type and IM Strains
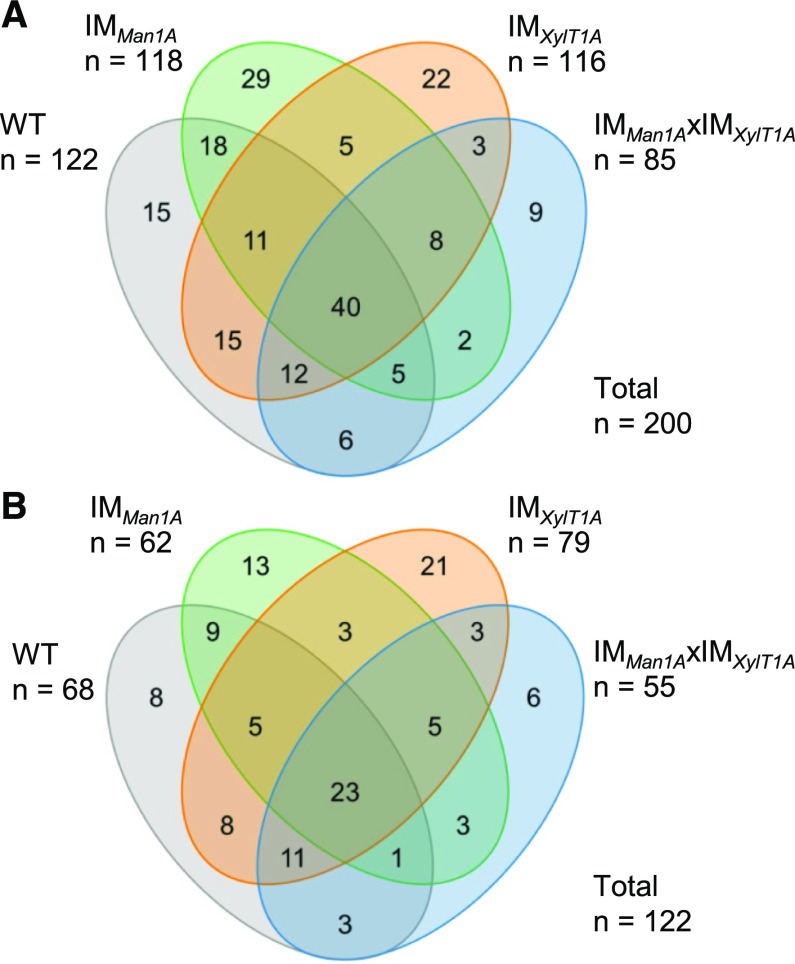
Taken together, the N-glycoproteomic characterization of the culture supernatant of all analyzed strains resulted in the identification of 181 different N-glycosites. For comparative purposes, results for progenies after crossing with CC-124 have been combined with their corresponding parental strain, that is IMMan1A and CC-124xIMMan1A Ins+, IMXylT1A and CC-124xIMXylT1A Ins+ as well as the wild type and Ins− strains, since their N-glycan phenotypes are identical. Of all identified N-glycosites, 27 have been found in all four strains (Fig. 5A). The N-glycan composition was assignable for a subset of 111 N-glycosites, whereby nine N-glycosites were common to all analyzed strains (Fig. 5B). Based on these common N-glycosites, different characteristics of N-glycans will be compared in the following.
Figure 5.
N-Glycosite identifications in all analyzed strains. A, Venn diagram for N-glycosites identified by database search engines. B, Venn diagram for N-glycosites for which the N-glycan composition could be determined. Results for progenies of genetic crossings with CC-124 have been included for the corresponding parental strain.
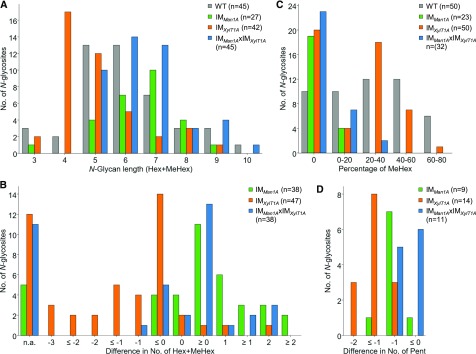
Direct comparison of N-glycan length, that is the number of Hex and methylated Hex residues attached to one N-glycosite, confirmed the presence of shorter N-glycans in IMXylT1A compared to all other strains (Fig. 6, A and B). In contrast, for N-glycans synthesized by IMMan1A and IMMan1AxIMXylT1A, a slight increase in N-glycan length in comparison to the wild type was observed. The degree of methylation in the wild type was nearly equally distributed ranging from 0 to 80%, while it was slightly lower in IMXylT1A (Fig. 6C). Strikingly, the broad majority of N-glycans from IMMan1A and IMMan1AxIMXylT1A was lacking MeHex and N-glycans that were still methylated showed only a low degree of methylation, which might be explained by residual Man1A activity. Considering N-glycosites that harbored N-glycans with two Pent in the wild type, IMMan1A and IMMan1AxIMXylT1A N-glycans were mostly lacking one Pent, whereas in IMXylT1A, N-glycans showed a decrease by at least one, often two Pent (Fig. 6D). N-Glycans modified with dHex were identified for all strains. However, this modification was observed less often for N-glycopeptides from IMMan1A when compared to N-glycosites harboring dHex in the wild type (Supplemental Fig. S6). This tendency for a lack of core dHex was less pronounced for IMMan1AxIMXylT1A and not observed for IMXylT1A N-glycopeptides.
Figure 6.
Comparison of N-glycan characteristics for wild type, IMMan1A, IMXylT1A, and IMMan1AxIMXylT1A. A, Comparison of N-glycan length, defined here as the sum of Hex and MeHex, (23 compared N-glycosites). B, Differences in N-glycan length (lengthIM − lengthWT) for N-glycosites identified in the wild type and respective IM strain. Some N-glycosites harboring multiple N-glycoforms could not be assigned to one of the categories (n.a.). C, Comparison of the percentage of MeHex in respect to N-glycan length (23 compared N-glycosites). D, Differences in the number of Pent (PentIM − PentWT) for N-glycosites that carried two Pent in the wild type. For each comparison, the legend indicates the total number of N-glycopeptides (A and C) and the total number of N-glycosites (B and D) for each strain, respectively. Results for progenies of genetic crossings with CC-124 have been included for the corresponding parental strain.
Immunoblot Analysis of IM Strains
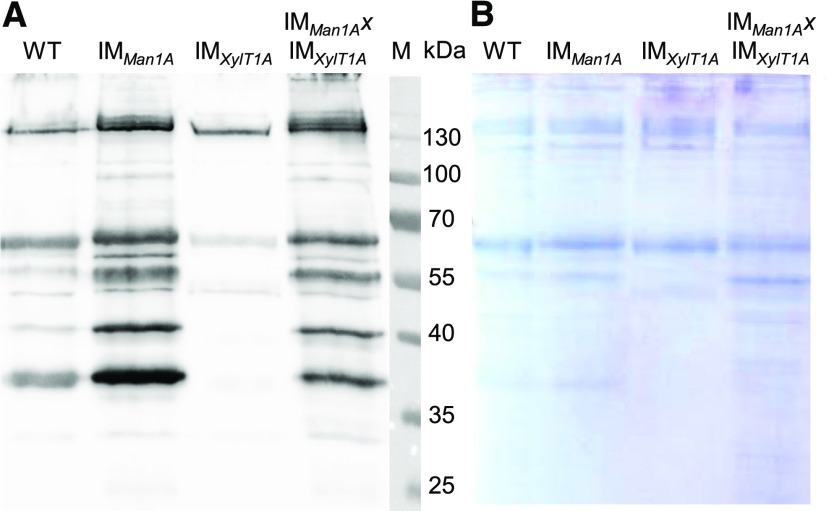
A lack of core Xyl was indicated for most N-glycans in IMXylT1A by MS and was verified in an immunoblot analysis after SDS-PAGE separation of whole cell extracts using an anti-Horseradish-Peroxidase (α-HRP) antibody, recognizing β1,2-Xyl and α1,3-Fuc residues (Fig. 7). Since the Coomassie-stained control gel confirmed equal loading of the supernatant (SN) samples, differences in signal intensity can be attributed to differential affinity of the α-HRP antibody. The weak signal for IMXylT1A in comparison to wild-type SN samples confirmed the lack of β1,2-core Xyl, since the remaining signal can be explained by binding of the antibody to Fuc epitopes. No signal decrease was observed for samples from IMMan1A and IMMan1AxIMXylT1A, suggesting the presence of core Xyl in both strains. Since the majority of mass spectrometrically analyzed N-glycopeptides in these strains were found to carry only one Pent, it can be concluded that they are lacking the terminal Xyl, which is linked in a β1,4-dependend manner. When comparing wild type and IMMan1A/IMMan1AxIMXylT1A, even an increase in signal intensity is visible. This might be explained by an altered accessibility of the core Xyl between N-glycans of the wild-type and mutant strains, possibly due to MeHex shielding the core Xyl or altering the N-glycan conformation, which has been shown to be important for detection by α-HRP (Kaulfürst-Soboll et al., 2011).
Figure 7.
Immunodetection of β1,2-Xyl residues in all analyzed strains. Proteins (9 µg) from the culture SN of the wild type, IMMan1A, IMXylT1A, and IMMan1AxIMXylT1A were separated by SDS-PAGE and either analyzed by western blot using α-HRP antibodies directed against β1,2-Xyl and α1,3-Fuc (A) or stained, as a loading control, using Coomassie Blue (B).
DISCUSSION
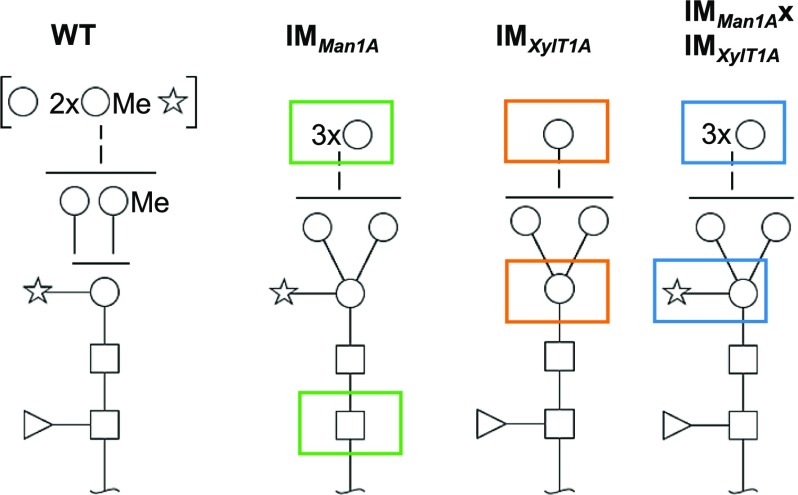
In this work, three C. reinhardtii strains carrying inserts in genes proposed to be involved in N-glycosylation, namely, man1A and xylT1A, were identified. The disturbed expression of Man1A and/or XylT1A resulted in distinct N-glycan compositions, deviating from the wild-type composition (Fig. 8). This implies different roles as well as a functional interconnection of the two enzymes in the N-glycosylation pathway of C. reinhardtii.
Figure 8.
Schematic representation of most common N-glycans identified in the wild type and respective IM strains. Disturbed expression of Man1A affects the modification of N-glycans with MeHex, terminal Pent and Fuc (green boxes), while insertional mutagenesis of XylT1A leads to shortened N-glycans lacking core Pent (orange boxes). N-Glycans from the double mutant show restored core Pent but a decrease in MeHex and terminal Pent in comparison to the wild type. Monosaccharides depicted above the solid horizontal line can be bound to any subjacent residue, to monosaccharides connected by a dashed line or to monosaccharides within the same bracket. Monosaccharide symbols follow the Symbol Nomenclature for Glycans (Varki, 2009).
In contrast to the wild type, IMXylT1A N-glycans were shown to be lacking core β1,2-Xyl by MS as well as immunoblot analyses. These results are in agreement with those obtained from single knockout mutants of XylT (AtXylT) in Arabidopsis, which also resulted in the absence of core Xyl attached to N-glycans (Strasser et al., 2004). Thus, it can be concluded that XylT1A functions as core β1,2-XylT in C. reinhardtii. In addition to the lack of core Xyl as an expected consequence of disturbed xylT1A expression, IMXylT1A N-glycans were mainly one to three Hex residues shorter than those found in the wild type. Since no N-glycans could be identified that were modified by two Xyl while, at the same time, being excessively trimmed, we propose a central role for the core Xyl in regulating N-glycan trimming in C. reinhardtii: After attachment of a core Xyl, no further trimming by mannosidases occurs. In addition to this inhibitory role in N-glycan trimming, CrXylT1A showed uncommon substrate specificity in comparison to XylTs of other organisms. For example, AtXylT activity is limited toward N-glycans harboring an additional β1,2-linked GlcNAc attached to the α1,3-linked core Man (Kajiura et al., 2012). Since C. reinhardtii does not encode for any GnTs, a different substrate specificity for CrXylT1A is apparent and in line with a low sequence identity of 23% with AtXylT (Mathieu-Rivet et al., 2013).
To define the role of Man1A, data obtained for IMMan1A as well as IMMan1AxIMXylT1A need to be taken into account. Based on the N-glycoproteomic analysis of IMMan1A, which revealed, in comparison to the wild type, a lack of MeHex and terminal Pent for most N-glycan compositions, a dependence of N-glycan methylation on Man1A activity can be postulated. While methylation of N-glycans is a rather uncommon modification in eukaryotes, it has been found in several algae, viz. Porphyridium sp. (Levy-Ontman et al., 2011), C. reinhardtii (Mathieu-Rivet et al., 2013), and Botryococcus braunii (Schulze et al., 2017). Additionally, N-glycan methylation has been reporter in archaea (Jarrell et al., 2014), molluscs, nematodes, planaria, and amoebae (Staudacher 2012; Schiller et al., 2012). Although the underlying mechanisms of N-glycan methylation remain unclear, S-adenosyl-Met (SAM)-dependent methyltransferases (MeTs) have been proposed to play a role in the glycosylation process of Haloferax volcanii and Caenorhabditis elegans (Magidovich et al., 2010; Wohlschlager et al., 2014). However, since Man1A belongs to the glycoside hydrolase family 47, containing α1,2-mannosidases, and is not expected to possess any SAM-dependent MeT domains, a direct involvement of the enzyme in the methylation of Hex residues is rather unlikely. Therefore, a multiple sequence alignment has been performed, employing Protein BLAST (Altschul et al., 2005), to identify potential MeTs in C. reinhardtii. Since no eukaryotic N-glycan MeTs are characterized so far, sequences of known archaeal N-glycan MeTs have been used for the search and yielded three candidates: Cre07.g346400, Cre06.g269250, and Cre09.g386747 (Supplemental Table S1). All three sequences contain a SAM-dependent MeT domain (Conserved Domain Database ID: cl17173) but have not been characterized so far.
For further insights in Man1A function, the N-glycan structures synthesized in IMMan1AxIMXylT1A should be considered. A role of Man1A as MeT would not affect the trimming observed for IMXylT1A. In the double mutant, however, the length of N-glycans is highly similar to those of IMMan1A, that is even slightly increased in comparison to the wild type. This strongly suggests a Man1A-dependent N-glycan trimming leading to N-glycans with one to three Hex residues less in IMXylT1A compared to wild type and the double mutant, thereby confirming hexosidase activity of Man1A.
In any case, the majority of N-glycans present in both IMMan1A and the double mutant is still trimmed. Hence, Man1A is not the only class I α-mannosidase involved in trimming of N-glycans in C. reinhardtii, which is in line with observations in other organisms (Liebminger et al., 2009; Herscovics 2001). In Arabidopsis, for example, three mannosidases (MNS1-3) are expressed, out of which MNS3 was shown to cleave off one Man residue from the Man9GlcNAc2 precursor in the ER. MNS1/2 on the other hand both need to be knocked out for suppressing the synthesis of complex type N-glycans, as they have redundant functions (Liebminger et al., 2009). In C. reinhardtii, the most promising candidate for a second α1,2- mannosidase is Cre06.g301600 (Man1B), annotated in the most recent release of the JGI database (version 5.5).
Another striking feature of N-glycans in IMMan1AxIMXylT1A is the presence of a core Xyl, which is in contrast to IMXylT1A. For both single mutant strains, disturbed gene expression and mutual inheritance of an altered N-glycan composition with the respective insertional cassette was confirmed. Since the double mutant lacks any detectable XylT1A on protein level (Supplemental Fig. S5E), the modification of N-glycans with β1,2-core XylT in IMMan1AxIMXylT1A indicates additional β1,2-core XylT activity independent of XylT1A. This XylT1B, which might be encoded in the genome as Cre16.g678997, showed no activity in IMXylT1A, that is in the presence of Man1A. Therefore, XylT1B activity is probably either inhibited by the increased N-glycan trimming in IMXylT1A or dependent on an altered N-glycan processing in IMMan1AxIMXylT1A as discussed below.
Furthermore, the transfer of a terminal Pent to decorated oligomannosidic N-glycans was strongly diminished in the absence of MeHex in IMMan1A, indicating that terminal xylosylation is favored by former Man1A-dependent trimming and methylation of the N-glycan. However, Man1A activity is not a prerequisite for terminal xylosylation, since N-glycans harboring two Pent were observed for IMMan1AxIMXylT1A. Similarly, although core fucosylation seems to be affected in IMMan1A, activity of the two putative fucosyltransferases (FucT, Cre18.g749697, Cre18.g749047) was not dependent on N-glycan methylation or Man1A activity. Surprisingly, the decrease in terminal xylosylation and core fucosylation was less prominent in IMMan1AxIMXylT1A. This could be explained by an accumulation or prolonged retention of immaturely N-glycosylated proteins in the Golgi apparatus due to the knockout of two N-glycosylation pathway enzymes. Thereby, even a low activity of XylT2 and FucT on Man1A-independent N-glycans might be sufficient to result in an increased addition of terminal Pent as well as core Fuc in IMMan1AxIMXylT1A in comparison to IMMan1A. Alternatively, an increased expression of XylT2 and FucT in the double mutant could have similar effects.
Altogether, our data indicate an enzymatic cascade in the N-glycosylation pathway of C. reinhardtii (Supplemental Fig. S7). In this process, XylT1A plays a key role in regulating N-glycan trimming, which, in turn, is accomplished in a Man1A-dependent manner and is favorable for methylation of Hex residues as well as the transfer of core Fuc and terminal Pent. However, the specificity of Man1A and additional mannosidases toward distinct N-glycan branches remains to be elucidated.
Although not all steps in the N-glycosylation pathway of C. reinhardtii are fully understood yet, the analyzed strains represent an important step toward the N-glycoengineering of C. reinhardtii for its potential use as an alternative platform for the production of biopharmaceuticals. Especially IMXylT1A, which shows only minor allergenic potential due to its lack of core Xyl, could be a basis for further development of strains useful for the production of biopharmaceuticals that require the presence of oligomannosidic N-glycans, like taliglucerase alpha (Brumshtein et al., 2010) or vascular endothelial growth factor (Claffey et al., 1995). However, further knockout of FucTs as well as the putative second core XylT might be required, particularly for IMMan1AxIMXylT1A, to achieve uniform N-glycosylation lacking nonhuman-like modifications.
Furthermore, N-glycoproteins identified here and previously (Mathieu-Rivet et al., 2013) comprise various flagellar proteins as well as secreted and cell surface proteins, several of them being involved in, for example, nutrient acquisition. Thus, it would be of high interest to assess the consequences of an altered N-glycan structure for the biology and in particular for acclimation responses of C. reinhardtii. However, since the functions of N-glycans can range from correct targeting to enzyme activity or recognition by receptors (Lannoo and van Damme, 2015; Varki, 2017), this is beyond the scope of this study. Nevertheless, the identified IM strains set the stage to analyze the physiological impact of altered and distinct N-glycan compositions in C. reinhardtii.
In this study, three C. reinhardtii strains differing in their N-glycan compositions were generated and analyzed. Taking those distinct changes in N-glycan compositions into account, the N-glycosylation pathway of this alga could be refined, especially in regard to the uncommon modification of N-glycans by MeHex and terminal Xyl as well as a novel regulatory mechanism of N-glycan trimming. Furthermore, the presented modifications of the N-glycosylation pathway in C. reinhardtii will shed new light on the functional importance of N-glycosylation for various N-glycoproteins in C. reinhardtii, including human homologs.
MATERIALS AND METHODS
Growth Conditions
If not indicated otherwise, all Chlamydomonas reinhardtii strains were grown photoheterotrophically in TAP medium at 25°C and 20 µE m−2 s−1, either as liquid cultures shaking at 120 rpm or on TAP plates containing 1.5% agar. For quantitative proteomics experiments, isotopic 14N and 15N labeling was performed using 14N and 15N ammonium chloride, respectively.
Insertional Mutagenesis and Identification of IM Strains
An insertional mutant library was generated and screened by PCR as described by Cheng et al. (2017) using a rescued mutant of ift46-1 (CC-4375) as the parental strain (wild type). Sequences for the insert specific primer LGR06-F as well as the gene specific primers used for screening are listed in Supplemental Table S2. The positive PCR products were sequenced using LGR06-F to identify the insertion site. After picking up single clones of IMMan1A and IMXylT1A, insertion of the AphVIII fragment was confirmed with a primer pair binding adjacently to the respective insertion site.
Analysis of man1A and xylT1A Transcript Levels Using RT-qPCR
Total RNA of Chlamydomonas cells was isolated using TRIzol reagent (Thermo Fisher Scientific). RNA concentration was determined by Quawell 5000 (Quawell Technology). Residual genomic DNA in total RNA was removed by DNase I (RNase-free) (Thermo Fisher Scientific). Reverse transcription of RNA was performed with oligo(dT) primers using First Strand cDNA Synthesis Kit (Thermo Fisher Scientific). Quantitative real-time PCR was performed using ABI 7900HT Fast Real-Time PCR System (Applied Biosystems) with SYBR Green Real-Time PCR Master Mix (TOYOBO) according to the manufacturer’s instructions. The housekeeping gene β-subunit-like polypeptide (CBLP, Cre06.g278222) was used as an internal control (Schloss 1990). The expression of ManI and XylT1A was quantified applying standard curve assay in SDS 2.4 software (Applied Biosystems) and normalized to CBLP. All primers used in real-time PCR are listed in Supplemental Table S2.
Quantification of Man1A and XylT1A Expression Levels by Parallel Reaction Monitoring
Cells were grown under LL conditions in 14N and 15N TAP media. The experiment was performed as label swap experiment. Cell amounts corresponding to 5 µg chlorophyll were mixed (IM strain and wild type) and pelleted by centrifugation (5,000g, 5 min). Cells were lysed in 100 mm Tris-HCl buffer, pH 7.6, containing 2% SDS, 1 mm PMSF, and 1 mm benzamidine using a sonication bath for 5 min. After removal of cell debris, cells were subjected to a Filter Aided Sample Preparation (FASP) protocol as described below using 0.35 µg trypsin.
The Q Exactive Plus was operated with the following PRM settings: resolution: 35,000 at m/z 200, AGC target: 1e5, maximum injection time: 400 ms, isolation window: 2.2 m/z. The gradient used for peptide elution with a flow rate of 300 nL min−1 is specified in Supplemental Table S3. Inclusion lists compiled with Skyline (MacLean et al., 2010; version 3.6) were used for scheduled fragmentation of target peptides.
The total peak areas of a minimum of three fragment ions per peptide were determined in Skyline with manual adjustment of peak borders. To correct for differences in total protein levels in the samples, peak areas were multiplied by a replicate-specific normalization factor derived from the mean abundance of three CF1 ATPase alpha subunit peptides. Peptide levels and standard deviations were calculated based on the peak areas of 14N and 15N labeled samples of the respective strain within the label swap. The .raw files have been uploaded together with the quantification results (see “Data Availability”).
Protein Isolation from the Culture Supernatant
The culture supernatant was obtained by three consecutive centrifugation steps. First, cells were pelleted at 2,000g for 3 min. Then the SN was centrifuged at 7,800g for 10 min and afterward at 48,000g for 2 h at 4°C. The resulting SN was concentrated by a factor of 100 by centrifugation in filter units (Amicon ultra centrifugal filters, 15 mL, 30 kD MWCO; Millipore). Protein concentration was determined by bicinchoninic acid assay (BCA Protein Assay Kit by Thermo Scientific Pierce). Samples were frozen in liquid nitrogen and stored at −80°C until use.
Immunoblot Analysis of N-Glycoproteins
Proteins from the SN were separated by SDS-PAGE (13% acrylamide) and either transferred to a nitrocellulose membrane or stained with Coomassie Blue G. Samples were loaded based on same protein amounts (9 µg). Membranes were blocked with 2% low-fat milk powder in PBS-T (3.2 mm Na2HPO4, 0.5 mm KH2PO4, 1.3 mm KCl, 135 mm NaCl, and 0.05% Tween, pH 7.4) for 16 h and then incubated with polyclonal rabbit antisera raised against HRP diluted 1:20,000 in PBS-T (α-HRP was kindly provided by Dr. Antje von Schaewen; Kaulfürst-Soboll et al., 2011) for 2 h. HRP-labeled anti-rabbit antibody (Bio-Rad) was used as secondary antibody 1:10,000 in PBS-T containing 2% low-fat milk powder for 1 h. Between each step, washing was performed three times with PBS-T. Western blots were developed by ECL and signals were digitally recorded with a Fusion-SL imaging system (Peqlab).
Crossing of IM Strains with CC-124 and Generation of IMMan1AxIMXylT1A Double Mutants
C. reinhardtii CC-124 (mt−) was mated with IMMan1A or IMXylT1A (mt+) to generate strains carrying the insert in a new genetic background as well as backcrossed IMMan1A or IMXylT1A, with wild-type man1A or xylT1A, respectively. Screening the progeny by colony PCR using the primer pair EX6-F-DNA, EX8-R-DNA as well as mating type specific primers resulted in the identification of CC-124xIMMan1A Ins− (mt+) and CC-124xIMMan1A Ins+ (mt−). Screening the respective progenies with primer pairs EX6-F, EX8-R, and LGR06, IN7-R as well as mating type-specific primers resulted in the identification of CC-124xIMXylT1A Ins− (mt+) and CC-124xIMXylT1A Ins+ (mt−).
C. reinhardtii strain IMXylT1A was mated with CC-124xIMMan1A Ins+ (mt−) to gain a strain carrying insertional cassettes in both genes. Screening of progenies by PCR with primer pairs for both loci as well as mating type primers resulted in identification of two double mutants with opposite mating types. Both strains had identical N-glycan patterns (data not shown). Only results for IMMan1AxIMXylT1A (mt+) are presented in this work.
FASP for N-Glycoproteomics
FASP was performed as previously described (Mathieu-Rivet et al., 2013; Wiśniewski et al., 2009) loading 30 µg to 60 µg protein from SN samples of wild-type and IM strains onto Amicon ultra centrifugal filters (0.5 mL, 30 kD MWCO; Millipore) and digesting with 0.3 to 0.6 µg trypsin (sequencing-grade modified, Promega) for 16 h at 37°C. Peptides were dried in a vacuum centrifuge and stored at −20°C. Samples from three biological replicates of each strain have been analyzed.
LC-MS Analysis
Peptides obtained from FASP were reconstituted in 2% (v/v) acetonitrile/0.1% (v/v) formic acid in ultrapure water and separated with an Ultimate 3000 RSLCnano System (Thermo Fisher Scientific). The sample was loaded on a trap column (C18 PepMap 100, 300 µm × 5 mm, 5-mm particle size, 100 Å pore size; Thermo Fisher Scientific) and desalted for 5 min using 0.05% (v/v) TFA/2% (v/v) acetonitrile in ultrapure water with a flow rate of 10 µL min−1. Peptides were then separated on a separation column (Acclaim PepMap100 C18, 75 mm i.d., 2-mm particle size, 100-Å pore size; Thermo Fisher Scientific) with a length of 50 cm (for wild type, IMXylT1A, and IMMan1AxIMXylT1A) or 15 cm (wild type, IMMan1A). The mobile phases were composed of 0.1% (v/v) formic acid in ultrapure water (A) and 80% acetonitrile/0.08% formic acid in ultrapure water (B). The gradient used for peptide elution with a flow rate of 300 nL min−1 is specified in Supplemental Table S3.
The LC system was coupled via a nanospray source to a Q Exactive Plus mass spectrometer (Thermo Fisher Scientific) operating in positive ion mode. MS data were acquired at a resolution of 70,000 for MS1. Fragmentation by higher-energy C-trap dissociation for MS2 (resolution of 17,500) was triggered in a data-dependent manner dynamically choosing the 12 most abundant precursor ions. Further details for the employed methods are listed in Supplemental Table S3. However, it should be noted that IS-CID was applied for the analysis of intact N-glycopeptides leading to the fragmentation of glycosidic bonds before MS1 (Mathieu-Rivet et al., 2013; Hsiao and Urlaub 2010). For these measurements, the 12 most abundant precursor ions were chosen by mass tags using masses of ±203.079373 (corresponding to the neutral loss of one HexNAc) with charges from 1 to 4 and 5 ppm mass tolerance.
Identification of Peptide Spectrum Matches and Statistical Postprocessing
Analysis of MS2 spectra was performed using the Python framework Ursgal (version 0.4.0; Kremer et al., 2016). Basically, the analysis pipeline consisted of a conversion of mzML to mgf files employing pymzML (Bald et al., 2012), search against a peptide database using X!Tandem (Craig and Beavis 2003), MS-GF+ (Kim et al., 2010), and OMSSA (Geer et al., 2004) and statistical postprocessing of unified results with qvality (Käll et al., 2009) for results obtained from total cell extracts and SN, respectively. The database contained protein sequences of the Chlamydomonas v5.5 gene models (Joint Genome Institute) merged with mitochondrial and chloroplast protein sequences from NCBI databases BK000554.2 and NC_001638.1 as well as sequences from the Common Repository of Adventitious Proteins (http://www.thegpm.org/crap/) resulting in a database with 18,941 proteins. Additionally, decoy sequences generated by peptide shuffling were included for all target proteins. In general, the default profile ‘QExactive+’ was used including a precursor ion tolerance of 5 ppm and a fragment ion tolerance of 20 ppm. When applied during sample preparation, carbamidomethylation of Cys was set as a fixed modification. Additionally, Met oxidation and N-terminal acetylation were included as potential posttranslational modifications for all samples. Furthermore, when IS-CID was applied, “HexNAc” and “HexNAc(2)” on Asn were added as optional modifications. Resulting peptide spectrum matches have been filtered by posterior error probability ≤1% and decoy hits were removed. For the identification of N-glycopeptides, peptide spectrum matches were filtered for peptides containing the N-glycosylation consensus sequence N-X-S/T. Further details on parameters can be gained from the Ursgal log JSONs that have been uploaded together with the mzML result files as well as the corresponding final Ursgal result files (see “Data Availability”).
Annotation of N-Glycan Compositions
The analysis of intact N-glycopeptides by IS-CID allows for the identification of the peptide sequence on MS2 level while the N-glycan composition can be deduced from a series of neutral losses in the corresponding MS1 spectra. This annotation of N-glycan compositions has been performed automatically using the in-house-developed Python tool SugarPy as described in Schulze et al. (2017). Briefly, SugarPy builds up all possible combinations for a list of defined monosaccharides and a maximal glycan length. Here, we have used the following monosaccharides: HexNAc (C8H13NO5), Hex (C6H10O5), MeHex (C7H12O5), dHex (C6H10O4), Pent (C5H8O4), and a maximal glycan length of 15. These combinations are added to glycopeptides identified using Ursgal (Kremer et al., 2016) and the resulting library of theoretical glycan tree-peptide combinations was matched on all MS1 spectra employing pyQms (Leufken et al., 2017) for accurate calculation and matching of isotope patterns. After validation based on mScore (≥0.5) and sub tree coverage (at least 70%), SugarPy scores were plotted against the retention time to define elution profiles of glycopeptides. For each glycopeptide elution peak a peptide identification (retention time ±1 min) was required to accept the identified glycan composition. Finally, spectra annotated by SugarPy, employing Plotly (Plotly Technologies collaborative data science; https://plot.ly), were reviewed manually for each assigned glycan composition. Those annotated spectra can be found in Supplemental Data S3.
Multiple Sequence Alignment for Potential MeTs
Since no eukaryotic N-glycan MeTs have been characterized so far, protein sequences from H. volcanii AglP and Halobacterium salinarumVNG_1065C have been used as queries against target protein databases from C. reinhardtii as well as Botryococcus braunii AC761 and CCALA778, for which N-glycans modified with MeHex have been observed as well (Schulze et al., 2017). Protein BLAST (Altschul et al., 2005) has been employed for the alignment and results were filtered for an E-value ≤ 1e-04. Identified sequences from B. braunii were used as queries in a second alignment step using the same target databases. Finally, all result sequences were subjected to a Conserved Domain Search (Marchler-Bauer and Bryant, 2014).
Data Availability
Mass spectrometry data have been uploaded to the ProteomeXchange Consortium via the PRIDE partner repository (Vizcáino et al., 2013) with the data set identifier PXD005254 for the analysis of intact N-glycopeptides experiment and PXD005257 for PRM experiments. All annotated spectra for the analysis of the N-glycan composition can be found in Supplemental Data S3.
Accession Numbers
Sequence data for genes/proteins studied in this article can be found in the Phytozome database (https://phytozome.jgi.doe.gov) under accession numbers Cre07.g336600 (man1A) and Cre09.g391282 (xylt1A).
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Identification of a man1A insertional mutant.
Supplemental Figure S2. N-glycoproteomic and genetic characterization of CC124xIMMan1A progenies.
Supplemental Figure S3. Identification of a xylT1A insertional mutant.
Supplemental Figure S4. N-Glycoproteomic and genetic characterization of CC124xIMXylT1A progenies.
Supplemental Figure S5. N-glycoproteomic and genetic characterization of IMMan1AxIMXylT1A progenies.
Supplemental Figure S6. Comparison of dHex modification of N-glycans from wild type, IMMan1A, IMXylT1A, and IMMan1AxIMXylT1A.
Supplemental Figure S7. Proposed model for the N-glycosylation pathway in C. reinhardtii.
Supplemental Table S1. Potential N-glycan MeTs identified by multiple sequence alignment.
Supplemental Table S2. List of primer sequences used for the screening and confirmation of IMMan1A, IMXylT1A, and IMMan1AxIMXylT1A.
Supplemental Table S3. List of MS related parameters that have been used for the analysis intact N-glycopeptides using IS-CID.
Supplemental Data S1. Fasta file containing the insert sequence as well as the sequencing results for the determination of the insertion sites in the analyzed mutant strains.
Supplemental Data S2. Summary of identified N-glycan compositions for all strains analyzed.
Supplemental Data S3. Annotated spectra for all identified N-glycopeptides are sorted by the corresponding strain.
Acknowledgments
M.H. acknowledges support from the Deutsche Forschungsgemeinschaft (HI 739/12-1) and by the Sino-German Center, Beijing, China (project GZ990). K.H. acknowledges support from National Natural Science Foundation of China (grant 31371354). G.L. acknowledges support from National Natural Science Foundation of China (grant 31400654).
Footnotes
M.H. acknowledges support from the Deutsche Forschungsgemeinschaft (HI 739/12-1) and by the Sino-German Center, Beijing, China (project GZ990). K.H. acknowledges support from National Natural Science Foundation of China (grant 31371354). G.L. acknowledges support from National Natural Science Foundation of China (grant 31400654).
These authors contributed equally to the article.
Articles can be viewed without a subscription.
References
- Altschul SF, Wootton JC, Gertz EM, Agarwala R, Morgulis A, Schäffer AA, Yu Y-K (2005) Protein database searches using compositionally adjusted substitution matrices. FEBS J 272: 5101–5109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bald T, Barth J, Niehues A, Specht M, Hippler M, Fufezan C (2012) pymzML--Python module for high-throughput bioinformatics on mass spectrometry data. Bioinformatics 28: 1052–1053 [DOI] [PubMed] [Google Scholar]
- Brumshtein B, Salinas P, Peterson B, Chan V, Silman I, Sussman JL, Savickas PJ, Robinson GS, Futerman AH (2010) Characterization of gene-activated human acid-beta-glucosidase: crystal structure, glycan composition, and internalization into macrophages. Glycobiology 20: 24–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng X, Liu G, Ke W, Zhao L, Lv B, Ma X, Xu N, Xia X, Deng X, Zheng C, et al. (2017) Building a multipurpose insertional mutant library for forward and reverse genetics in Chlamydomonas. Plant Methods 13: 36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claffey KP, Senger DR, Spiegelman BM (1995) Structural requirements for dimerization, glycosylation, secretion, and biological function of VPF/VEGF. Biochim Biophys Acta 1246: 1–9 [DOI] [PubMed] [Google Scholar]
- Craig R, Beavis RC (2003) A method for reducing the time required to match protein sequences with tandem mass spectra. Rapid Commun Mass Spectrom 17: 2310–2316 [DOI] [PubMed] [Google Scholar]
- Fanata WID, Lee KH, Son BH, Yoo JY, Harmoko R, Ko KS, Ramasamy NK, Kim KH, Oh D-B, Jung HS, et al. (2013) N-glycan maturation is crucial for cytokinin-mediated development and cellulose synthesis in Oryza sativa. Plant J 73: 966–979 [DOI] [PubMed] [Google Scholar]
- Geer LY, Markey SP, Kowalak JA, Wagner L, Xu M, Maynard DM, Yang X, Shi W, Bryant SH (2004) Open mass spectrometry search algorithm. J Proteome Res 3: 958–964 [DOI] [PubMed] [Google Scholar]
- Harris EH, Stern DB, Witman GB, eds (2009) The Chlamydomonas Sourcebook, Ed 2. Academic Press, London [Google Scholar]
- Herscovics A. (2001) Structure and function of Class I α 1,2-mannosidases involved in glycoprotein synthesis and endoplasmic reticulum quality control. Biochimie 83: 757–762 [DOI] [PubMed] [Google Scholar]
- Hochmal AK, Zinzius K, Charoenwattanasatien R, Gäbelein P, Mutoh R, Tanaka H, Schulze S, Liu G, Scholz M, Nordhues A, et al. (2016) Calredoxin represents a novel type of calcium-dependent sensor-responder connected to redox regulation in the chloroplast. Nat Commun 7: 11847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsiao H-H, Urlaub H (2010) Pseudo-neutral-loss scan for selective detection of phosphopeptides and N-glycopeptides using liquid chromatography coupled with a hybrid linear ion-trap/orbitrap mass spectrometer. Proteomics 10: 3916–3921 [DOI] [PubMed] [Google Scholar]
- Jarrell KF, Ding Y, Meyer BH, Albers S-V, Kaminski L, Eichler J (2014) N-linked glycosylation in Archaea: a structural, functional, and genetic analysis. Microbiol Mol Biol Rev 78: 304–341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kajiura H, Okamoto T, Misaki R, Matsuura Y, Fujiyama K (2012) Arabidopsis β1,2-xylosyltransferase: substrate specificity and participation in the plant-specific N-glycosylation pathway. J Biosci Bioeng 113: 48–54 [DOI] [PubMed] [Google Scholar]
- Käll L, Storey JD, Noble WS (2009) QVALITY: non-parametric estimation of q-values and posterior error probabilities. Bioinformatics 25: 964–966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang JS, Frank J, Kang CH, Kajiura H, Vikram M, Ueda A, Kim S, Bahk JD, Triplett B, Fujiyama K, et al. (2008) Salt tolerance of Arabidopsis thaliana requires maturation of N-glycosylated proteins in the Golgi apparatus. Proc Natl Acad Sci USA 105: 5933–5938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaulfürst-Soboll H, Rips S, Koiwa H, Kajiura H, Fujiyama K, von Schaewen A (2011) Reduced immunogenicity of Arabidopsis hgl1 mutant N-glycans caused by altered accessibility of xylose and core fucose epitopes. J Biol Chem 286: 22955–22964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S, Mischerikow N, Bandeira N, Navarro JD, Wich L, Mohammed S, Heck AJR, Pevzner PA (2010) The generating function of CID, ETD, and CID/ETD pairs of tandem mass spectra: applications to database search. Mol Cell Proteomics 9: 2840–2852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kremer LPM, Leufken J, Oyunchimeg P, Schulze S, Fufezan C (2016) Ursgal, universal Python module combining common bottom-up proteomics tools for large-scale analysis. J Proteome Res 15: 788–794 [DOI] [PubMed] [Google Scholar]
- Lannoo N, van Damme EJM (2015) Review/N-glycans: the making of a varied toolbox. Plant Sci 239: 67–83. [DOI] [PubMed] [Google Scholar]
- Leufken J, Niehues A, Sarin P, Wessel F, Hippler M, Leidel SA, Fufezan C (2017) pyQms enables universal and accurate quantification of mass spectrometry data. Mol Cell Proteomics 16: 1736–1745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy-Ontman O, Arad SM, Harvey DJ, Parsons TB, Fairbanks A, Tekoah Y (2011) Unique N-glycan moieties of the 66-kDa cell wall glycoprotein from the red microalga Porphyridium sp. J Biol Chem 286: 21340–21352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liebminger E, Hüttner S, Vavra U, Fischl R, Schoberer J, Grass J, Blaukopf C, Seifert GJ, Altmann F, Mach L, et al. (2009) Class I alpha-mannosidases are required for N-glycan processing and root development in Arabidopsis thaliana. Plant Cell 21: 3850–3867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacLean B, Tomazela DM, Shulman N, Chambers M, Finney GL, Frewen B, Kern R, Tabb DL, Liebler DC, MacCoss MJ (2010) Skyline: an open source document editor for creating and analyzing targeted proteomics experiments. Bioinformatics 26: 966–968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magidovich H, Yurist-Doutsch S, Konrad Z, Ventura VV, Dell A, Hitchen PG, Eichler J (2010) AglP is a S-adenosyl-L-methionine-dependent methyltransferase that participates in the N-glycosylation pathway of Haloferax volcanii. Mol Microbiol 76: 190–199 [DOI] [PubMed] [Google Scholar]
- Marchler-Bauer A, Bryant SH (2004) CD-Search: protein domain annotations on the fly. Nucleic Acids Res 32: W327–331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathieu-Rivet E, Scholz M, Arias C, Dardelle F, Schulze S, Le Mauff F, Teo G, Hochmal AK, Blanco-Rivero A, Loutelier-Bourhis C, et al. (2013) Exploring the N-glycosylation pathway in Chlamydomonas reinhardtii unravels novel complex structures. Mol Cell Proteomics 12: 3160–3183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merchant SS, Prochnik SE, Vallon O, Harris EH, Karpowicz SJ, Witman GB, Terry A, Salamov A, Fritz-Laylin LK, Marechal-Drouard L, et al. (2007) The Chlamydomonas genome reveals the evolution of key animal and plant functions. Science 318: 245–250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiller B, Hykollari A, Yan S, Paschinger K, Wilson IBH (2012) Complicated N-linked glycans in simple organisms. Biol Chem 393: 661–673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schloss JA. (1990) A Chlamydomonas gene encodes a G protein beta subunit-like polypeptide. Mol Gen Genet 221: 443–452 [DOI] [PubMed] [Google Scholar]
- Schulze S, Urzica E, Reijnders MJMF, van de Geest H, Warris S, Bakker LV, Fufezan C, Martins Dos Santos VAP, Schaap PJ, Peters SA, et al. (2017) Identification of methylated GnTI-dependent N-glycans in Botryococcus brauni. New Phytol 215: 1361–1369 [DOI] [PubMed] [Google Scholar]
- Staudacher E. (2012) Methylation--an uncommon modification of glycans. Biol Chem 393: 675–685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strasser R. (2016) Plant protein glycosylation. Glycobiology 26: 926–939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strasser R, Altmann F, Mach L, Glössl J, Steinkellner H (2004) Generation of Arabidopsis thaliana plants with complex N-glycans lacking beta1,2-linked xylose and core alpha1,3-linked fucose. FEBS Lett 561: 132–136 [DOI] [PubMed] [Google Scholar]
- Strasser R, Bondili JS, Vavra U, Schoberer J, Svoboda B, Glössl J, Léonard R, Stadlmann J, Altmann F, Steinkellner H, et al. (2007) A unique β1,3-galactosyltransferase is indispensable for the biosynthesis of N-glycans containing Lewis a structures in Arabidopsis thaliana. Plant Cell 19: 2278–2292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strasser R, Stadlmann J, Schähs M, Stiegler G, Quendler H, Mach L, Glössl J, Weterings K, Pabst M, Steinkellner H (2008) Generation of glyco-engineered Nicotiana benthamiana for the production of monoclonal antibodies with a homogeneous human-like N-glycan structure. Plant Biotechnol J 6: 392–402 [DOI] [PubMed] [Google Scholar]
- Vanier G, Lucas P-L, Loutelier-Bourhis C, Vanier J, Plasson C, Walet-Balieu M-L, Tchi-Song PC, Remy-Jouet I, Richard V, Bernard S, et al. (2017) Heterologous expression of the N-acetylglucosaminyltransferase I dictates a reinvestigation of the N-glycosylation pathway in Chlamydomonas reinhardtii. Sci Rep 7: 10156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varki A, ed (2009) Essentials of Glycobiology, Ed. 2 Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY: [PubMed] [Google Scholar]
- Varki A. (2017) Biological roles of glycans. Glycobiology 27: 3–49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiśniewski JR, Zougman A, Nagaraj N, Mann M (2009) Universal sample preparation method for proteome analysis. Nat Methods 6: 359–362 [DOI] [PubMed] [Google Scholar]
- Vizcaíno JA, Côté RG, Csordas A, Dianes JA, Fabregat A, Foster JM, Griss J, Alpi E, Birim M, Contell J, et al. (2013) The PRoteomics IDEntifications (PRIDE) database and associated tools: status in 2013. Nucleic Acids Res 41: D1063–D1069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wohlschlager T, Butschi A, Grassi P, Sutov G, Gauss R, Hauck D, Schmieder SS, Knobel M, Titz A, Dell A, et al. (2014) Methylated glycans as conserved targets of animal and fungal innate defense. Proc Natl Acad Sci USA 111: E2787–E2796 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Mass spectrometry data have been uploaded to the ProteomeXchange Consortium via the PRIDE partner repository (Vizcáino et al., 2013) with the data set identifier PXD005254 for the analysis of intact N-glycopeptides experiment and PXD005257 for PRM experiments. All annotated spectra for the analysis of the N-glycan composition can be found in Supplemental Data S3.