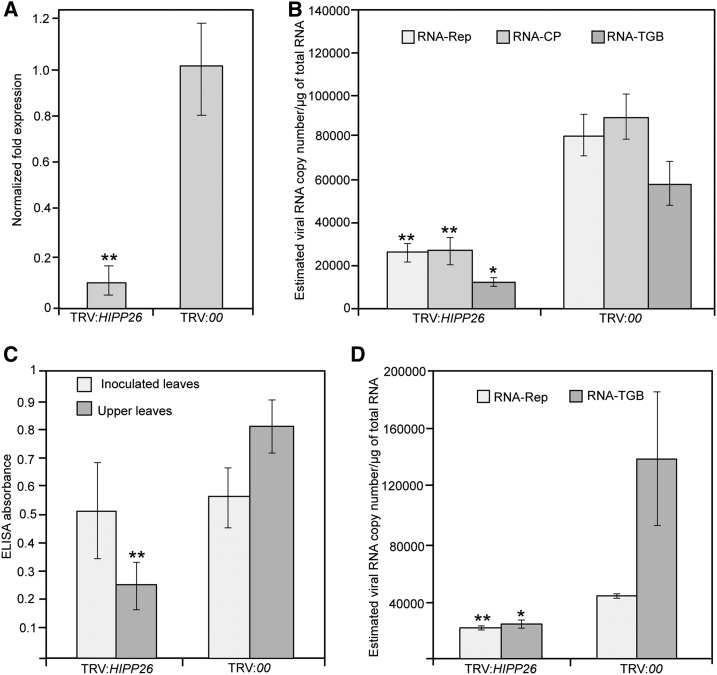
Figure 9.
Effects of the knockdown of NbHIPP26 expression on PMTV accumulation in upper noninoculated leaves. A, Gene silencing of the NbHIPP26 gene in TRV:HIPP26-infected plants. Total RNAs were extracted from the upper leaves 14 dpi and were used for RT-qPCR. Data are means ± sd from three experiments (n = 14 for each treatment). NbF-BOX and NbPP2A were used as normalization controls. B, Effect of HIPP26 gene silencing on viral RNA levels in upper noninoculated leaves. Total RNAs were extracted from the upper leaves 14 dpi with PMTV and used for absolute RT-qPCR. Ten-fold serial dilutions of full-length infectious cDNA clones of RNA-Rep, RNA-CP, and RNA-TGB were used to obtain standard curves and determine viral RNA copy number. NbF-BOX and NbPP2A were used as normalization controls. Data are means ± sd from four independent experiments with three or four biological replicates each. C, ELISA was used to determine PMTV infection in inoculated and upper noninoculated leaves of treated plants. Absorbance values (indicating the presence of PMTV capsid protein) in samples of inoculated leaves were similar in both treatments but were more than 50% greater in the upper noninoculated leaves of plants inoculated with empty vector, showing suppression of the long-distance movement of PMTV particles in NbHIPP26 knockdown plants. D, RT-qPCR quantification of RNA-Rep and RNA-TGB in upper noninoculated leaves of plants challenged with these two PMTV genomic components. Data are means ± sd; n = 6, two independent experiments. Asterisks indicate statistically significant differences compared with TRV:00: *, P < 0.01 and **, P ≤ 0.05, by Student’s two-tailed t test.