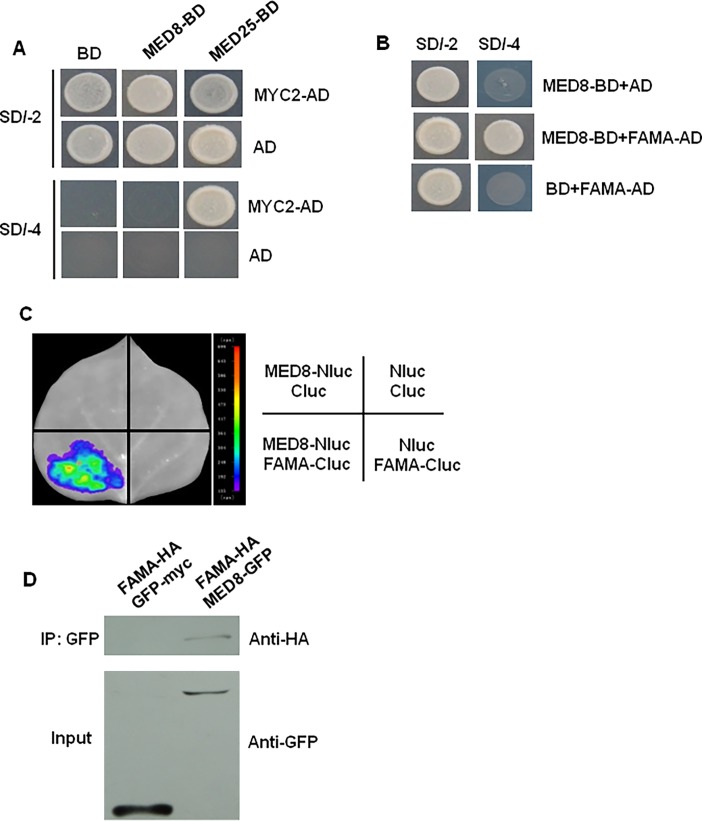
Fig 2. MED8 can interact with FAMA, but not with MYC2.
(A) A Y2H assay was used to detect the interactions of MED8 with the MYC2 protein. Yeast cells co-transformed with pGADT7-MYC2 (preys) and pGBKT7-MED8 (baits) were selected and subsequently grown on yeast synthetic dropout lacking Leu and Trp (SD/-2) as a transformation control, or on selective media lacking Ade, His, Leu, and Trp (SD/-4) to test protein interactions. The pGADT7-MYC2 (preys) and pGBKT7-MED25 (baits) interaction constituted a positive control. pGADT7-MYC2 co-transformed with the pGBDT7 vector, and pGBKT7-MED8 or pGBKT7-MED25 co-transformed with the pGADT7 vector were included as controls. (B) A Y2H assay used to detect the interactions of MED8 with the FAMA protein. Yeast cells co-transformed with pGADT7-FAMA (preys) and pGBKT7-MED8 (baits) were selected and subsequently grown on yeast synthetic dropout lacking Leu and Trp (SD/-2) as a transformation control, or on selective media lacking Ade, His, Leu, and Trp (SD/-4) to test protein interactions. pGADT7-FAMA co-transformed with the pGBDT7 vector, and pGBKT7-MED8 co-transformed with the pGADT7 vector were included as controls. (C) Split-luc assays showing that MED8 can interact with FAMA in N. benthamiana leaves. Three biological replicates were performed, and similar results were obtained. (D) Co-IP assays were used to verify the interaction of MED8 with FAMA in N. benthamiana leaves. Protein extracts from N. benthamiana leaves infiltration with both 35Spro:FAMA-HA and 35Spro:MED8-GFP (FAMA-HA MED8-GFP) or 35Spro:GFP-myc (FAMA-HA GFP-myc) was immunoprecipitated (IP) with the GFP antibody, and immunoprecipitated proteins were analyzed by immunoblotting using anti-HA and anti-GFP antibodies. The experiments were repeated three times, with similar results.