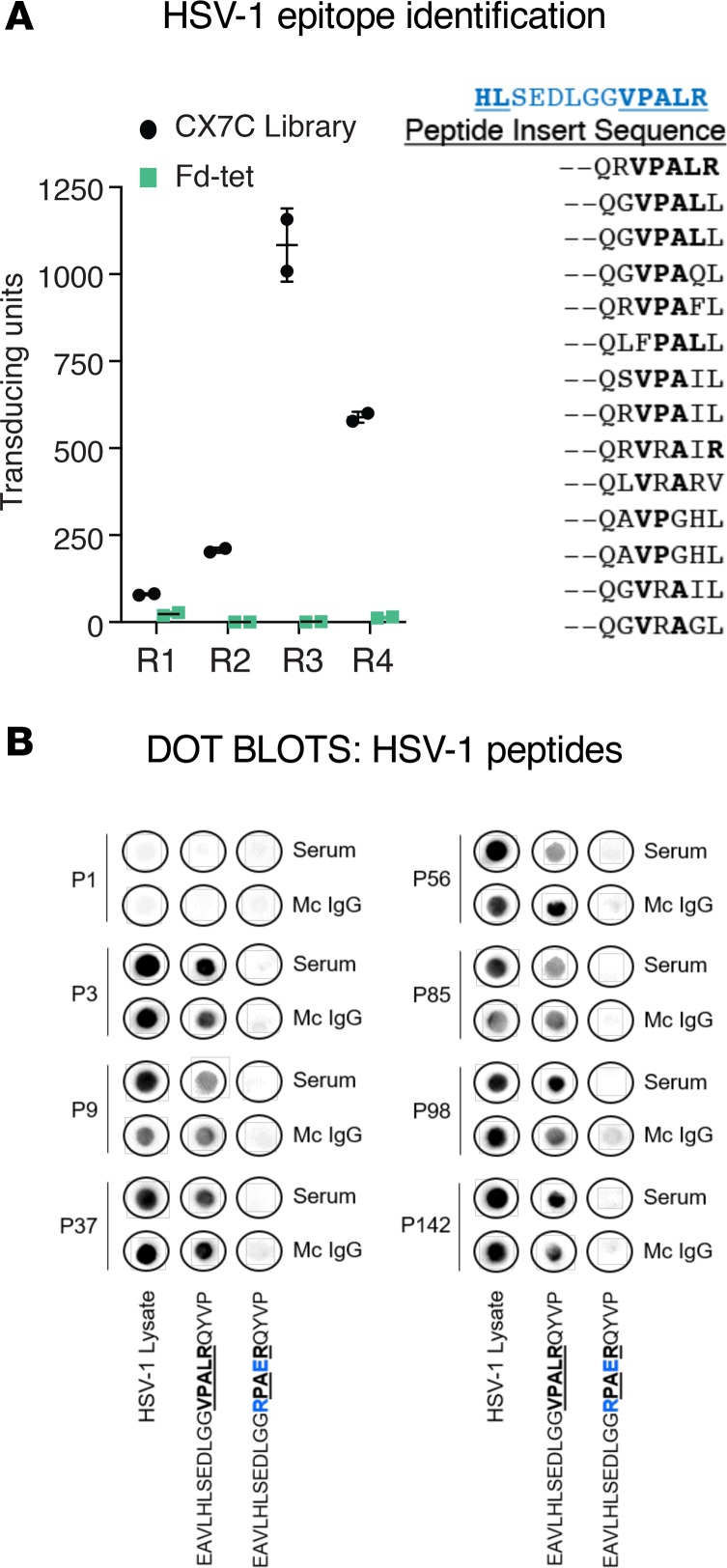
Figure 11. Confirmation of the specificity of HSV-1 recognition of purified mc IgGs.
(A, left): The purified monoclonal (mc) IgG from herpes simplex virus-1–positive (HSV-1+) index patient P56 was immobilized on Protein A/G beads and used for 4 rounds (R1–R4) of phage display peptide purification, with a CX7C library; the Fd-tet lacking peptide inserts served as a negative control. After the fourth round of selection, bacterial colonies were picked at random for sequencing. Results are expressed as mean ± SEM of duplicate wells. (A, right) Alignment of 14 related sequences, used for a BLAST alignment identifying the sequence VPALR from HSV-1 tegument protein, UL36 (in blue, aa 1498–1510). (B) Dot blotting with HSV-1 lysate and relevant (EAVLHLSEDLGGVPALRQYVP) and irrelevant (EAVLHLSEDLGGRPAERQYVP) HSV-1–derived peptides was performed for serum and purified mc IgG from the 7 patients found to have HSV-1–specific mc IgG with the multiplex infectious-antigen array (MIAA) assay. Patient P1, whose mc IgG did not recognize HSV-1 using the MIAA assay, was used as a negative control. Amino acids of the relevant epitope are in bold and underlined. The modified amino acids of the irrelevant peptide are in blue. Dot blots were performed at least twice.