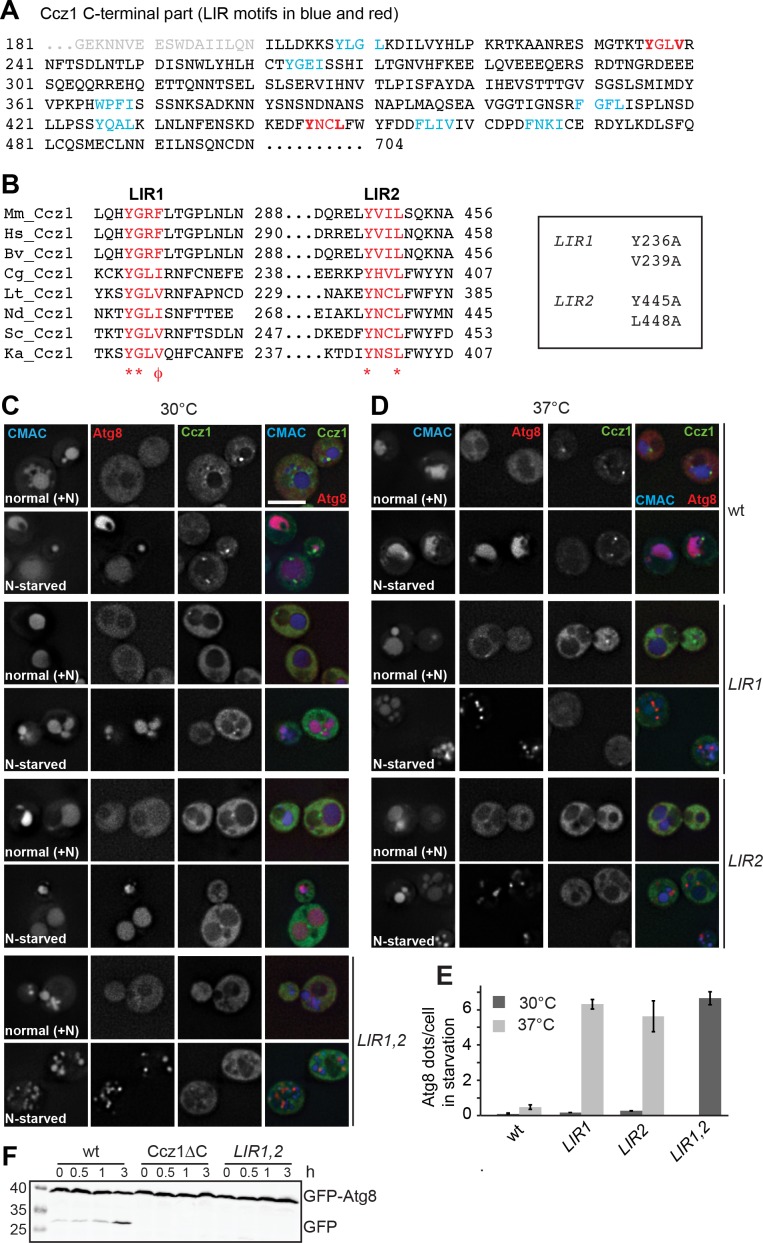
Figure 3. Identification of the LIR motifs in Ccz1 required for function.
(A) Schematic representation of potential LIR motifs of the C-terminal part of Ccz1. Blue and red indicates all LIR motifs analyzed, red the motifs that also impair Ccz1 localization. (B) Alignments of the potential Ccz1 LIR motifs Mm: Mus musculus, Hs: Homo sapiens, Cg: Candida glabrata, Lt: Lachancea thermotolerans, Nd: Naumovozyma dairenensis, Ka: Kazachstania Africana. (C–D) Effect of LIR mutants on localization, autophagy and vacuole morphology. Analysis was done as in Figure 1B–H. CMAC staining was done for 15 min before analysis. Cells were grown either at 30°C or 37°C during growth or starvation. Size bar, 5 µm. (E) Quantification of Atg8 dots per cell from images in (C–D). Error bars represent SD. (F) Analysis of autophagy over time. Cells were grown at 30°C and incubated in starvation medium for the indicated time periods, then harvested, and proteins were analyzed by SDS-PAGE and Western blotting against GFP.