Figure 3.
HIC1 Deficiency Alters iTreg Cell Transcriptome and Positions the Cells for Alternative Fates
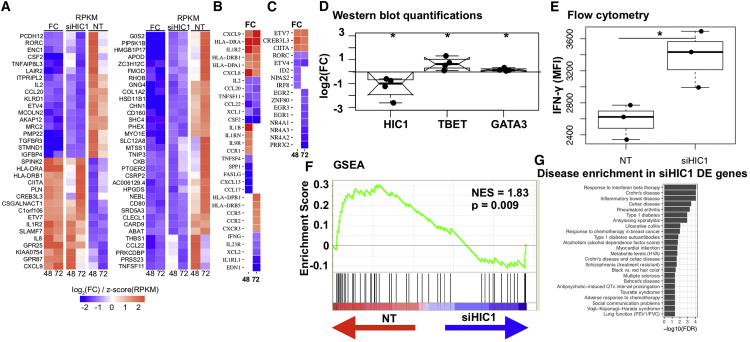
(A–C) Heatmaps showing the expression of top genes DE (FDR < 0.05, |log2[FC]| > 1) upon HIC1 silencing. siHIC1 and NT columns show expression (RPKM) in HIC1-deficient and HIC1-sufficient conditions, respectively. FC columns show fold change (iTreg/Th0) of the expression: (A) top DE genes (FDR = 0.05, |log2[FC]| > 1) that were common to the two time points; (B) cytokines and chemokines and their receptors at 48 and 72 hr; (C) TFs at 48 and 72 hr.
(D) NT- or siHIC1-treated cells were cultured in iTreg condition for 3 days, followed by WB. Blots were quantitated in ImageJ (NIH). Normalization was done by GAPDH. Log2(FC) was calculated with respect to NT samples. Significance was determined using two-tailed paired t test. ∗p < 0.05. Data from four biological replicates are shown.
(E) Boxplot showing IFN-γ expression in NT- or siHIC1-treated iTreg cells when reactivated after 72 hr with PMA and ionomycin. Data from three biological replicates are shown. Significance was determined using two-tailed paired t test. ∗p < 0.05.
(F) iTreg signature genes were top upregulated genes (log2FC > 2) in iTreg conditions compared with Th0 at 48 hr (Table S1). Gene set enrichment analysis (GSEA) (Subramanian et al., 2005) was then used to test where the genes from this set lie in the ranked HIC1 knockdown (KD) 48 hr data. Each vertical line in the plot shows one gene. Lines toward the red indicate genes that are enriched in NT sample, and those toward the blue indicate enrichment in siHIC1 samples.
(G) Bar chart showing the NHGRI diseases and traits whose SNPs are enriched around (±100 kb) genes DE upon HIC1 silencing. All the diseases are enriched at an FDR of 0.05, and those that cross the dotted line are enriched at an FDR of 0.01.