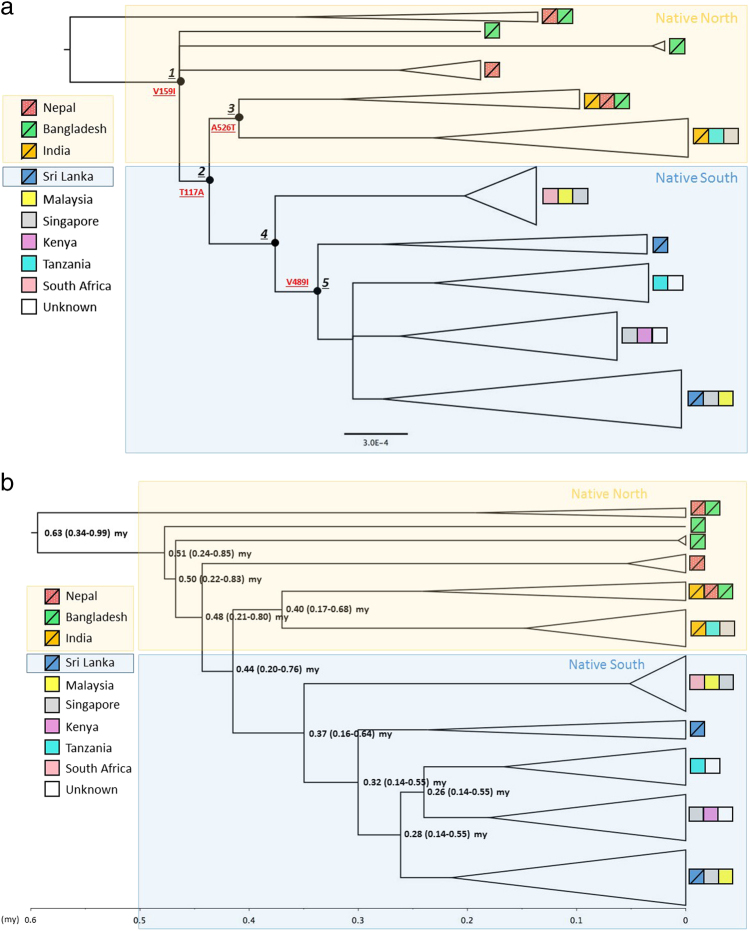
Fig. 2.
a Rooted phylogenetic summary tree (50% majority rule) estimated using Bayesian inference in MrBayes. All non-collapsed nodes were well supported (PP > 0.9). Main Clades are numbered and indicated with black circles. The candidate variants under positive selection: V159I in ND2 and T117A, A526T and V489I in ND5 gene indicated for relevant branches. The outgroup (not shown) was Corvus macrorhynchos. b BEAST dated phylogenetic tree where numbers show mean (and 95% highest posterior densities) estimates of divergene time between lineages (in million years). Both trees inferred based on 13 protein-coding genes representing House Crow lineages originating from different locations; terminal tips colour coded based on the geographical origin of individuals. Native range indicated by squares with diagonal lines. Very short branches within species clades have been collapsed for better presentation; triangle height corresponds to number of sequences and triangle width corresponds to nucleotide diversity within clade