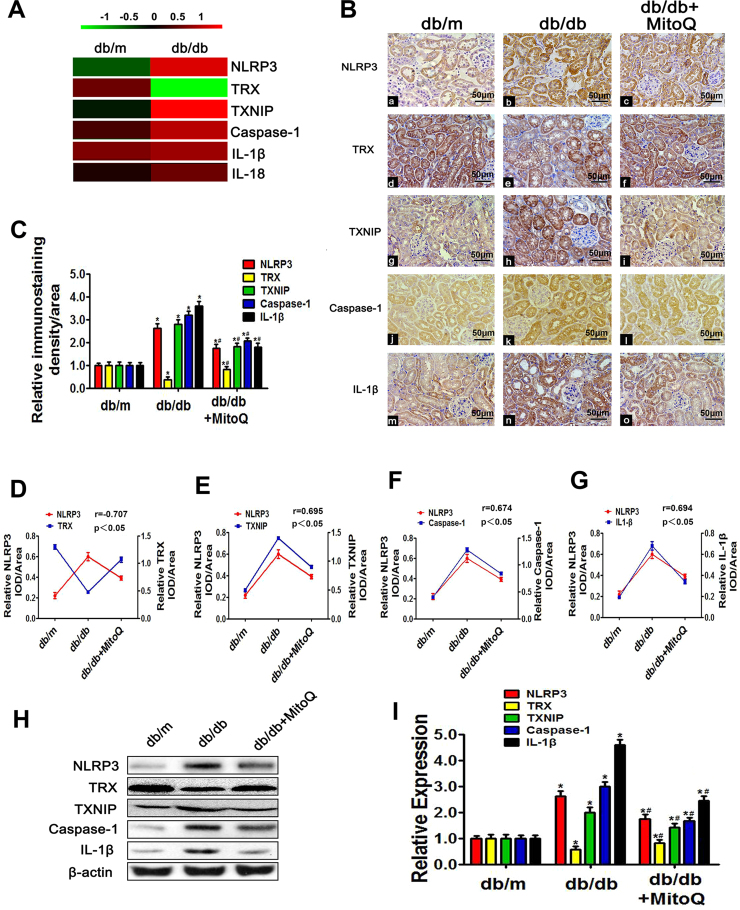
Fig. 4.
Effects of MitoQ on renal NLRP3, TRX, TXNIP, Caspase-1 and IL-1β expression in db/db mice.A: Renal expression of the proteins detected by iTRAQ-based quantitative proteomics analysis in db/m and db/db mice. Green represents down-regulation, red represents up-regulation, and black represents no change. The bar code (− 1–1) at the top shows the color scale of the log2 values. B: Immunohistochemical analysis of NLRP3 (Ba-c), TRX (Bd-f), TXNIP (Bg-i), Caspase-1 (Bj-l) and IL-1β expression (Bm-o) in kidney tissues. (magnification × 200). C: Quantification of NLRP3, TRX, TXNIP, Caspase-1 and IL-1β expression in tissues from the different groups. D-G: Analysis of the correlations between NLRP3 expression and TRX, TXNIP, Caspase-1 and IL-1β expression: r (NLRP3/TRX) = −0.707 (D), r (NLRP3/TXNIP) = 0.695 (E), r (NLRP3/Caspase-1) = 0.674 (F) and r (NLRP3/IL-1β) = 0.694 (G). H: Western blotting analysis of NLRP3 (H, first line), TRX (H, second line), TXNIP (H, third line), Caspase-1 (H, fourth line) and IL-1β expression (H, last line) in the kidneys in the different groups of mice. I: Densitometric analysis of the WB analysis results. Values are the mean ± SE; r: correlation coefficient; *P < 0.05 vs control group; #P < 0.05 vs db/db group. n = 12.