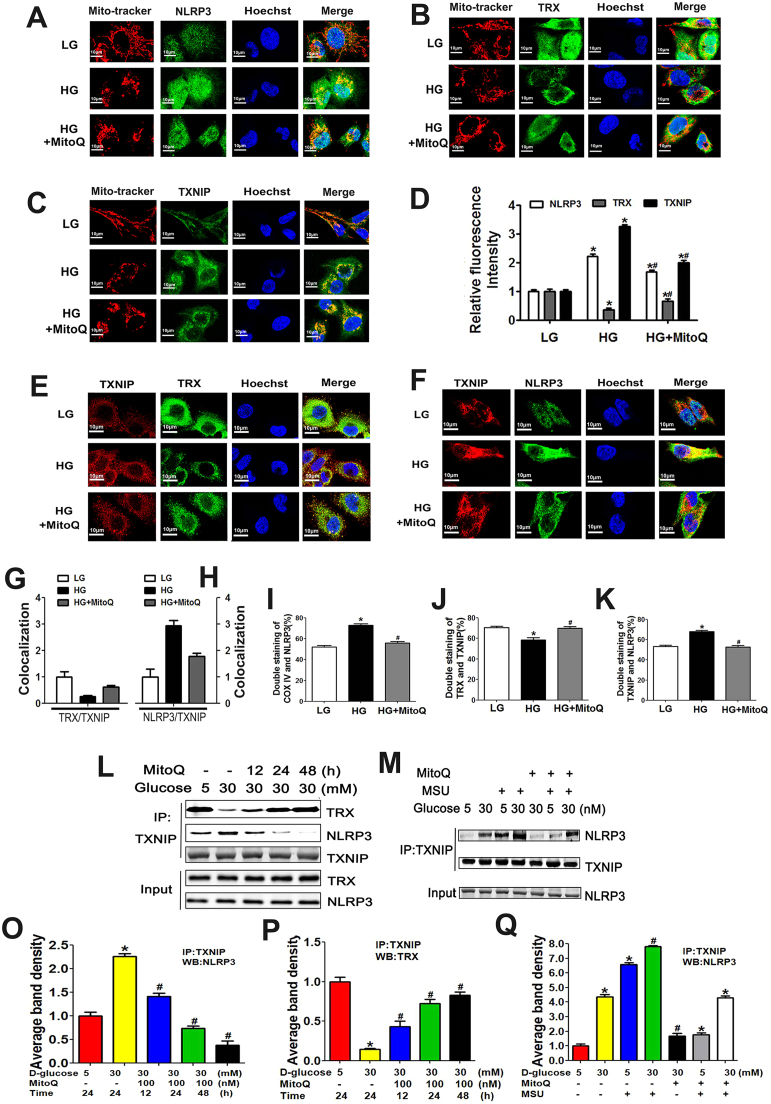
Fig. 7.
Effects of MitoQ on the associations between NLRP3 expression and TRX and TXNIP expression in HK-2 cells subjected to an HG environment. A-D: IF analysis showing MMP and NLRP3 (A), TRX (B), and TXNIP expression (C) in HK-2 cells exposed to HG conditions and pretreated with or without MitoQ. D: Quantification of NLRP3, TRX and TXNIP expression in HK-2 cells exposed to HG and pretreated with or without MitoQ. E-H: Confocal immunofluorescence images and semi-quantification showing the co-localization of TRX and TXNIP (E, G) and TXNIP and NLRP3 (F, H) in HK-2 cells exposed to HG conditions and pretreated with or without MitoQ. I-K: Flow cytometry and quantification analysis of COX IV and NLRP3 expression (I), TRX-TXNIP interactions (J), and TXNIP-NLRP3 interactions (K) in HK-2 cells cultured under HG conditions and pretreated with or without MitoQ. L, M: Co-IP and WB analysis showing the interactions between TXNIP and NLRP3 and between TXNIP and TRX in HK-2 cells subjected to HG conditions and treated with MitoQ or MSU. N-P: Quantification analysis of the interactions between TXNIP and NLRP3 (N, P) and between TXNIP and TRX (O) in HK-2 cells cultured under HG conditions following MitoQ or MSU administration. Values are the mean ± SE; *P < 0.05 vs control group; #P < 0.05 vs HG group. n = 3.