Abstract
Background
Although asymptomatic carriage of Streptococcus pneumoniae (Spn) is common, acquisition of the bacteria is the first step in disease pathogenesis. We examined the effect of introduction of the 7-valent pneumococcal vaccine on Spn carriage patterns in a cohort of Peruvian children.
Methods
We used data from a prospective cohort study that collected monthly nasopharyngeal samples from children under 3 years of age. Spn isolates were serotyped using Quellung reactions, and bacterial density was determined by quantitative polymerase chain reaction. Changes in Spn carriage patterns, including the rate of carriage and number and density of serotypes carried over time, were evaluated before (2009) and after widespread vaccination with PCV7 (2011). Using all pneumococcal detections from each child and year, we identified serotypes that were present both at first and last detection as “persisters” and serotypes that replaced a different earlier type and were detected last as “recolonizers.”
Results
Ninety-two percent (467/506) of children in 2009 and 89% (451/509) in 2011 carried Spn at least once. In 2009 and 2011, rates of carriage were 9.03 and 9.04 Spn detections per person-year, respectively. In 2009, 23F, a serotype included in PCV7, was the only type identified as a persister and 6A, 15B, and 19A were identified as recolonizer serotypes. In 2011, 6B and 7C were persister serotypes and 13 was a frequent recolonizer serotype.
Conclusions
Overall Spn carriage among children under 3 in Peru was similar before and after introduction of PCV7; however, serotype-specific rates and longitudinal carriage patterns have shifted.
Keywords: asymptomatic carriage, pneumococcal carriage, pneumococcal colonization, pneumococcal conjugate vaccine, pneumococcal disease, Streptococcus, pneumoniae
Streptococcus pneumoniae is a common cause of severe bacterial pneumonia in young children, and can also cause otitis media, bacteremia, and meningitis. The World Health Organization estimates that pneumococcal disease accounted for 5% of the approximately 476 000 deaths that occurred worldwide in HIV-negative children under age 5 in 2008. The burden of pneumococcal disease is highly skewed toward populations of low socioeconomic status: greater than 90% of those deaths occurred among children from low-income countries [1–4]. In Peru, pneumococcal infections were estimated to account for 12 000–18 000 deaths annually prior to pneumococcal vaccine introduction [5]. A recent study of children hospitalized with invasive pneumococcal disease in Lima, Peru, estimated that 68% of all cases and 80% of fatal cases are among children younger than 2 years of age, underscoring the susceptibility of this age group to poor outcomes [6].
Although the vast majority of nasopharyngeal S. pneumoniae (Spn) colonization is asymptomatic, acquisition of the bacteria is the critical first step in disease pathogenesis. The highest prevalence of Spn colonization is among children, particularly children under two years of age [7, 8], though exact estimates of colonization prevalence among healthy children vary widely and depend heavily on a range of host and environmental factors [8, 9]. Children commonly carry multiple strains simultaneously as well as undergo serial loss and acquisition of strains over time [10, 11]. Colonized children are responsible for much of the horizontal spread of Spn serotypes, and as a result play an outsized role in the overall epidemiology of Spn colonization and disease [12]. Therefore, understanding carriage dynamics in this group is important for understanding the burden and dissemination of Spn in communities.
Introduction of a pneumococcal conjugate vaccine has multiple effects on the population-level dynamics of Spn colonization and carriage. Serotype replacement, the process whereby nonvaccine serotypes “move in” to the ecologic niche left vacant by vaccine serotypes, is well characterized in the context of pneumococcal vaccine introduction [13]. However, less is known about the effects of vaccine introduction on the dynamic process of carriage, including the order and frequency of Spn colonization by serotype.
In Peru, the heptavalent pneumococcal vaccine (PCV7), which protects against serotypes 4, 6B, 9V, 14, 18C, 19F, and 23F, was introduced into the national immunization program in 2009. The RESPIRA-PERU cohort collected information on pneumococcal carriage in young children from 2009 through 2011 [14]. A previous analysis on a subset of this cohort showed that overall carriage of vaccine types was reduced from 2009 to 2011 but did not examine more detailed patterns in carriage [15]. We examined the overall and serotype-specific rates of Spn carriage in the RESPIRA-PERU cohort and assessed changes in Spn carriage over time, before (in 2009) and after PCV7 introduction (in 2011), to determine whether certain serotypes were more likely than others to persist in children over time. Last, we explored serotype-specific colonization density as a potential explanation for changes in carriage patterns.
METHODS
Study Cohort
The cohort for this study was derived from a household-based prospective cohort study (RESPIRA-PERU) of young children conducted in the province of San Marcos, Cajamarca, Peru [14]. Briefly, field workers identified households with children younger than 3 years of age, obtained informed consent from parents, and administered questionnaires that collected baseline demographic and socioeconomic information from each household. Children were enrolled starting in May 2009 and followed until 3 years of age, loss to follow-up, or the end of the study (September 2011), whichever came first. To maintain a constant cohort size of approximately 500 children, children leaving the cohort were replaced by newborns in the study area. Median age (IQR) at cohort enrollment was 4.6 (0.5–17.1) months, 48% of children were female, and median duration in the study was 14.5 months [14].
We examined the subset of this cohort that was observed from May to November 2009 and May through September 2011, allowing for comparison of population-level Spn carriage patterns before and after widespread PCV7 use. PCV7 vaccination status of study participants was ascertained via questionnaire and verification of vaccination cards. Of note, PCV7 was progressively introduced in the study area. In 2009, 2% of enrolled children had received PCV7; in 2011, 70% of enrolled children were vaccinated.
Nasopharyngeal Sample Collection
Nasopharyngeal (NP) samples were collected monthly from children according to World Health Organization (WHO)–recommended procedures. Samples were collected with a Rayon polyester swab and were immediately placed in a 2.0-mL cryogenic tube with 1 mL of transport medium (comprised of skim milk–tryptone–glucose–glycerine [STGG]) [16]. Specimens were transported with cold packs to the local laboratory within 8 hours of collection and preserved with the swab in the medium at –70°C [17].
DNA Extraction and Quantitative Polymerase Chain Reaction
DNA extraction, quantitative polymerase chain reaction (qPCR), and serotyping by Quellung were performed using methods previously described [18, 19]. Pneumococcal density was quantified by qPCR reactions using the pan-pneumococcus lytA assay developed by the Streptococcus Laboratory at the Centers for Disease Control and Prevention [20]. A standard curve of lytA was generated with known DNA concentrations and plotted against the cycle threshold (CT) to yield the copy number, calculated as 10(CT − 33.701)/−3.4262) [21, 22].
Isolation and Identification of S. pneumoniae Strains
Nasopharyngeal swabs were thawed and vortexed, and 200 μL of the specimen was transferred to a 5-mL Todd-Hewitt broth containing 0.5% of yeast extract and 1 mL of rabbit serum (Gibco by Life Technologies, Carlsbad, CA) [23]. This enriched culture was incubated for 6 hours at 37°C, inoculated onto blood agar plates (tryptic soy agar plates supplemented with 5% sheep blood), and incubated for 18–24 hours at 37°C. The predominant strain was isolated by selecting a single colony from those that were most abundant and morphologically similar in culture, and identified using the optochin test (Remel, Lenexa, KS) and bile solubility test [23]. Initial S. pneumoniae serotyping was performed by multiplex PCR [24], and further serotype discrimination was carried out using Quellung reactions.
Changes in Colonization and Distribution of Colonizing Serotypes After PCV7
We compared colonizing serotypes in 2009 (pre-PCV7) and 2011 (post-PCV7). For each year, we calculated rates of colonization by dividing the number of Spn detections using culture by the sum total of person-time, in months, contributed by all children in that year. We considered each Spn detection as a separate carriage event, and carriage of a particular serotype did not preclude a new detection, or carriage, of that same type or a different type the next time a child was sampled. To assess whether the overall diversity of circulating serotypes had changed, we determined whether the number of distinct serotypes detected in each year was different in 2009 vs 2011, considering the total number of person-months contributed by children in each year. We calculated serotype-specific carriage rates for each “common” serotype (a “common” serotype was one detected in at least 20 swabs, or >1% of all NP swabs in that year). For serotypes common in both 2009 and 2011, we calculated rate ratios comparing serotype-specific carriage rates in each year.
For each child, we calculated the number of detections of Spn and the number of distinct serotypes detected during the study period. We calculated the mean duration of carriage for both years, defined as the period of time during which a child was continuously colonized by Spn (starting with the first Spn detection and over the time period that subsequent consecutive samples were positive for Spn). Importantly, this calculation assumes that children carried Spn continuously in between monthly NP swab collections. We compared these measures between the 2009 and 2011 cohorts.
Serotype-Specific Colonization and Density Patterns
Among children who had at least 2 positive detections, we identified which serotypes were detected first and last. We identified those serotypes that “persisted”: serotypes that were present at both first and last detection; and serotypes that “recolonized”: serotypes that displaced a different, earlier serotype and were detected last. We calculated persistence for each serotype by dividing the number of children in whom that serotype was observed at first and last detection by the total number of children with the serotype at first detection. We defined a persistent serotype as one with a persistence measure greater than 50%: in other words, it was detected both first and last in more than 50% of the children in which it was detected first. Recolonization was calculated by dividing the number of children with a different serotype at first and last detection by the total number of children with the serotype at last detection; a recolonizing serotype was one that displaced a different serotype in more than 50% of the children in which it was detected last. Last, we assessed differences in bacterial density between serotypes in the same year and within serotypes across years using the Mann-Whitney test.
RESULTS
Study Cohort
We analyzed samples from 506 children in 2009 and 509 children in 2011; the median ages at cohort entry were 14.1 months and 0.80 months, respectively. There were 1790 NP swabs (median 2 per child) collected from children in 2009 and 1803 (median 2 per child) in 2011. Spn was detected in 61% (1083/1790) of NP swabs collected in 2009 (the sum total of all samples from all children in 2009) and 53% (963/1803) of NP swabs collected in 2011 (P < .001). The total amounts of person-time contributed by children were 1434.7 person-months in 2009 and 1270.15 person-months in 2011; these were calculated by summing the total number of days from collection of the first to last NP swab for each child. The median time from first to last NP swab were 71 days in 2009 and 91 days in 2011 (Table 1). Median time of carriage, defined as the period of time during which a child was continuously colonized by Spn, were 63 days in 2009 and 56 days in 2011.
Table 1.
Characteristics of the RESPIRA-PERU Cohort in 2009 and 2011
| Characteristic | 2009 Cohort (n = 506) | 2011 Cohorta (n = 509) | P Value |
|---|---|---|---|
| Median age at cohort entry (IQR), mo | 14.1 (6.3–24.4) | 0.8 (0.3–4.6) | <.001 |
| Male sex, % | 53 | 50 | .34 |
| Total number of NP swabs collected | 1790 | 1803 | - |
| Number of NP swabs positive for Spn by culture | 1083 (61) | 963 (53) | <.001 |
| Median number of NP swabs per child (IQR) | 2 (1–4) | 2 (1–3) | .003 |
| Median time from first to last NP swab (IQR), d | 71 (49–140) | 91 (56–91) | .003 |
| Children with at least 1 swab positive for Spn by culture | 431 (85) | 415 (82) | .20 |
| Children with at least 1 swab positive for Spn by qPCR | 467 (92) | 451 (89) | .10 |
| Proportion of children with at least 1 swab Spn+ (culture or qPCR) | 467 (92) | 451 (89) | .10 |
| Person-time contributed by cohort children, person-months | 1434.7 | 1270.15 | - |
Abbreviations: IQR, interquartile range; NP, nasopharyngeal; qPCR, quantitative polymerase chain reaction.
aThe 2011 cohort included 182 children that joined the cohort during 2009.
Changes in Colonization and Distribution of Colonizing Serotypes After PCV7
Rates of colonization were similar in children before (2009) and after (2011) PCV7 introduction: 0.755 and 0.758 Spn detections per person-month (equivalent to 9.03 and 9.04 detections per person-year), respectively (Table 2).
Table 2.
Overall Number of Carriage Events and Distinct Serotype Carriage Events
| 2009 Cohort (n = 506) No. (%) |
2011 Cohort (n = 509) No. (%) |
|
|---|---|---|
| Overall number of carriage events (NP swabs positive for Spn), per child (%) | ||
| 0 | 75 (14.8) | 94 (18.5) |
| 1 | 129 (25.5) | 133 (26.1) |
| 2 | 116 (22.9) | 107 (21.0) |
| 3 | 89 (17.6) | 93 (18.3) |
| 4 | 50 (9.9) | 74 (14.5) |
| 5 | 29 (5.7) | 8 (1.6) |
| 6 | 17 (3.4) | - |
| 7 | 1 (<0.1) | - |
| Total carriage events | 1083 | 963 |
| Median | 2 | 2 |
| Rate, detections/person-month | 0.755 | 0.758 |
| Rate ratio (95% CI), reference: 2009 | 1.00 (0.970–1.14) | |
| P value for rate ratio | .22 | |
| Number of distinct serotypes detected, per child (%) | ||
| 0 | 75 (14.8) | 94 (18.5) |
| 1 | 201 (39.7) | 235 (46.2) |
| 2 | 139 (27.5) | 136 (26.7) |
| 3 | 60 (11.9) | 39 (7.7) |
| 4 | 24 (4.74) | 5 (1.0) |
| 5 | 7 (1.4) | - |
| Total carriage events of unique serotypes | 790 | 644 |
| Median | 1 | 1 |
| Rate, detections/person-month | 0.551 | 0.507 |
| Rate ratio (95% CI), reference: 2009 | 0.92 (0.91–1.09) | |
| P value for rate ratio | .94 | |
Abbreviations: CI, confidence interval; NP, nasopharyngeal.
The proportions of children with at least 1 detection of Spn by qPCR were 92% (467/506) in 2009 and 89% (451/509) in 2011. In both 2009 and 2011, the median number of positive NP swabs (Spn detections) per child was 2 (rate ratio, 1.00; 95% confidence interval [CI], 0.97–1.14) (Table 2).
During follow-up, 45% of children (230/506) in 2009 and 35% (180/509) in 2011 had more than 1 distinct serotype detected; 18% (91/506) of children in 2009 and 9% (44/509) in 2011 had more than 2 distinct serotypes detected (Table 2). The median number of distinct serotypes detected per child was 1 in both 2009 and 2011 (rate ratio, 0.92; 95% CI, 0.91–1.09).
Overall, common serotypes accounted for 72% of all detections in 2009 and 76% in 2011. Serotype-specific carriage rates of common types ranged from 0.0139 to 0.0773 detections/person-year in 2009, with PCV7 serotypes 23F and 6B showing the highest carriage rates. In 2011, serotype-specific rates ranged from 0.0213 to 0.0803 detections/person-year, with PCV7 type 19F and serotype 11A showing the highest rates of carriage (Table 3). The number of common non-PCV7 serotypes increased from 8 in 2009 to 11 in 2011, though this was not significant (P = .34). Carriage rates of non-PCV7 serotypes 6C (rate ratio, 1.30; 95% CI, 0.94–1.82), 10A (rate ratio, 3.88; 95% CI, 2.38–6.30); 11A (rate ratio, 3.30; 95% CI, 2.24–4.84), and 19A (rate ratio, 1.40; 95% CI, 1.11–1.65) increased in 2011 as compared with 2009. The carriage rates of PCV7 serotypes 6B (rate ratio, 0.53; 95% CI, 0.38–0.74) and 23F (rate ratio, 0.35; 95% CI, 0.80–1.14) were significantly reduced in 2011 as compared with 2009, though PCV7 serotype 19F (rate ratio, 1.19; 95% CI, 0.90–1.59) showed a modest, though not significant, increase in carriage rate (Table 3). There was a slightly greater diversity of serotypes observed in 2009 (54) as compared with 2011 (46), although this difference was not statistically significant (P = .85).
Table 3.
Distribution and Carriage Rates of Most Common Serotypes, Before and After PCV7
| Serotype | 2009 | 2011 | Rate Ratiob (95% CI), Ref: 2009 | ||
|---|---|---|---|---|---|
| Frequency | Rate, Person-Months | Frequency | Rate, Person-Months | ||
| 6A | 49 | 0.0342 | - | - | |
| 6Ba | 111 | 0.0773 | 52 | 0.0409 | 0.53 (0.38–0.74) |
| 6C | 65 | 0.0453 | 75 | 0.0590 | 1.30 (0.94–1.82) |
| 7C | - | - | 27 | 0.0213 | |
| 10A | 21 | 0.0146 | 72 | 0.0567 | 3.88 (2.38–6.30) |
| 11A | 35 | 0.0244 | 102 | 0.0803 | 3.30 (2.24–4.84) |
| 13 | - | - | 39 | 0.0307 | |
| 14a | 40 | 0.0278 | - | - | |
| 15A | - | - | 34 | 0.0268 | |
| 15B | 23 | 0.0160 | 26 | 0.0205 | 1.00 (0.57–1.75) |
| 15C | - | - | 38 | 0.0299 | |
| 17F | 20 | 0.0139 | - | - | |
| 19A | 29 | 0.0202 | 36 | 0.0283 | 1.40 (1.11–1.65) |
| 19Fa | 91 | 0.0634 | 96 | 0.0756 | 1.19 (0.90–1.59) |
| 23A | 21 | 0.0146 | - | - | |
| 23B | - | - | 42 | 0.0331 | |
| 23Fa | 110 | 0.0767 | 34 | 0.0268 | 0.35 (0.80–1.14) |
| 35F | - | - | 27 | 0.0213 | |
| NT | 119 | 0.0829 | 35 | 0.0276 | 0.33 (0.23–0.48) |
| Total | 738 | 735 | |||
Abbreviation: CI, confidence interval.
aPCV7 types.
bRate ratios only calculated for serotypes common in both years.
Serotype-Specific Colonization and Density Patterns
Among children with Spn detected in 2 or more samples, 55% of children in 2009 and 45% in 2011 had a different serotype detected at first and last NP swab. In 2009, the only persistent serotype was PCV7 type 23F. Recolonizing serotypes that displaced earlier, different serotypes included 6A, 6B, 6C, 10A, 11A, 14, 15B, 19A, and 19F. The most frequent recolonizing types (>70%) in 2009 were 6A, 15B, and 19A (Table 4A). In 2011, the persistent serotypes were 6B, 7C, and 11A. The recolonizing serotypes were 6C, 13, 15A, 15C, 19A, 19F, 23B, 23F, and 35F; the most common recolonizer (>70%) was serotype 13 (Table 4B).
Table 4A.
Most Common First and Last Serotypes Among Children With at Least 2 Serotypes in 2009 (n = 309)
| Last Serotype | 6A | 6Ba | 6C | 10A | 11A | 14a | 15B | 17F | 19A | 19Fa | 23A | 23Fa | NT | All Other Serotypes | Total |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| First serotype | |||||||||||||||
| 6A | 5 | 1 | 0 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 1 | 10 |
| 6Ba | 0 | 11 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 1 | 5 | 20 |
| 6C | 0 | 0 | 8 | 0 | 0 | 0 | 1 | 0 | 0 | 1 | 0 | 0 | 1 | 3 | 14 |
| 10A | 1 | 0 | 1 | 2 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 6 |
| 11A | 0 | 1 | 0 | 0 | 5 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 7 |
| 14a | 1 | 0 | 0 | 0 | 0 | 4 | 0 | 0 | 0 | 0 | 0 | 0 | 3 | 3 | 11 |
| 15B | 0 | 0 | 0 | 0 | 0 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 2 | 5 |
| 17F | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 3 | 0 | 0 | 0 | 0 | 0 | 1 | 5 |
| 19A | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 2 | 3 | 10 |
| 19Fa | 0 | 0 | 2 | 0 | 1 | 0 | 0 | 1 | 1 | 10 | 1 | 0 | 4 | 4 | 24 |
| 23A | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 4 | 0 | 0 | 2 | 8 |
| 23Fa | 2 | 2 | 1 | 0 | 1 | 1 | 0 | 0 | 0 | 2 | 0 | 16 | 1 | 5 | 31 |
| NT | 1 | 5 | 4 | 1 | 0 | 0 | 1 | 1 | 3 | 8 | 0 | 5 | 7 | 10 | 46 |
| All other serotypes | 6 | 11 | 7 | 2 | 3 | 3 | 6 | 0 | 3 | 6 | 3 | 7 | 8 | 41 | 106 |
| Total | 17 | 33 | 25 | 6 | 12 | 11 | 9 | 6 | 8 | 29 | 8 | 30 | 28 | 80 | 303 |
| Persistence, % | 29.4 | 33.3 | 32.0 | 33.3 | 41.7 | 36.4 | 11.1 | 50.0 | 12.5 | 34.5 | 50.0 | 53.3 | 25.0 | ||
| Recolonization, % | 70.6 | 66.7 | 68.0 | 66.7 | 58.3 | 63.6 | 88.9 | 50.0 | 88.5 | 65.5 | 50.0 | 46.7 | 75.0 | ||
a PCV7 types.
Table 4B.
Most Common First and Last Serotypes Among Children With at Least 2 Serotypes in 2011 (n = 281)
| Last Serotype | 6Ba | 6C | 7C | 10A | 11A | 13 | 15A | 15B | 15C | 19A | 19Fa | 23B | 23Fa | 35F | NT | All Other Serotypes | Total |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| First serotype | |||||||||||||||||
| 6Ba | 9 | 1 | 0 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 3 | 16 |
| 6C | 1 | 9 | 0 | 1 | 1 | 2 | 0 | 1 | 3 | 0 | 2 | 1 | 0 | 1 | 1 | 1 | 24 |
| 7C | 0 | 0 | 6 | 0 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 1 | 1 | 0 | 0 | 0 | 10 |
| 10A | 0 | 0 | 1 | 10 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 1 | 0 | 0 | 0 | 2 | 16 |
| 11A | 0 | 1 | 0 | 1 | 18 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 1 | 0 | 0 | 24 |
| 13 | 0 | 0 | 0 | 0 | 0 | 3 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 3 | 8 |
| 15A | 0 | 1 | 0 | 0 | 2 | 0 | 4 | 0 | 0 | 0 | 2 | 0 | 0 | 1 | 0 | 1 | 11 |
| 15B | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 3 | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 7 |
| 15C | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 4 | 0 | 1 | 0 | 0 | 0 | 1 | 1 | 8 |
| 19A | 0 | 0 | 1 | 0 | 2 | 0 | 0 | 0 | 0 | 4 | 0 | 0 | 0 | 0 | 0 | 2 | 9 |
| 19Fa | 1 | 1 | 0 | 0 | 2 | 2 | 0 | 0 | 0 | 3 | 12 | 0 | 1 | 1 | 2 | 2 | 27 |
| 23B | 0 | 0 | 0 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 3 | 5 | 0 | 0 | 0 | 2 | 13 |
| 23Fa | 1 | 0 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 2 | 2 | 3 | 0 | 1 | 1 | 13 |
| 35F | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 3 | 0 | 0 | 5 |
| NT | 0 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | 2 | 2 | 0 | 0 | 1 | 2 | 10 |
| All other serotypes | 2 | 7 | 1 | 4 | 5 | 3 | 3 | 2 | 1 | 4 | 2 | 3 | 1 | 1 | 9 | 32 | 80 |
| Total | 14 | 21 | 10 | 20 | 35 | 12 | 10 | 6 | 13 | 12 | 28 | 16 | 7 | 9 | 15 | 53 | 281 |
| Persistence, % | 64.3 | 42.9 | 60.0 | 50.0 | 51.4 | 25.0 | 40.0 | 50.0 | 30.8 | 33.3 | 42.9 | 31.3 | 42.9 | 33.3 | 6.7 | ||
| Recolonization, % | 35.7 | 57.1 | 40.0 | 50.0 | 48.6 | 75.0 | 60.0 | 50.0 | 69.2 | 66.7 | 57.1 | 68.8 | 57.1 | 66.7 | 93.3 | ||
a PCV7 types.
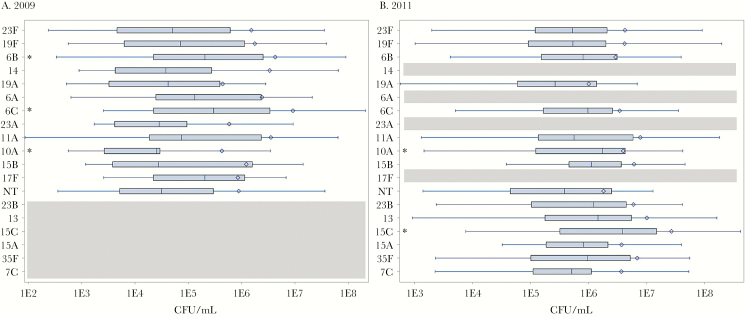
There were few differences in densities between serotypes within the same year; however, densities of almost all serotypes in 2011 were significantly higher than densities in 2009 (Supplementary Table 1). In 2009, NP samples containing 6B (P = .008) and 6C (P = .003) had slightly higher median densities when compared with the median density of all serotypes combined. In 2011, NP samples with predominant serotypes 10A (P = .041) and 15C (P = .002) had slightly higher median densities (Figure 1).
Figure 1.
Densities of the most common serotypes in 2009 and 2011. Boxplots for each serotype show the median and interquartile range, and diamonds indicate the mean, for the density of serotypes in each year. Asterisks indicate a density significantly different (P < .05) than the median density of all serotypes in that year. Serotypes are aligned for comparison across years; gray areas reflect serotypes that were not common in that year. Abbreviation: CFU, colony forming units.
DISCUSSION
We found similar overall rates of Spn carriage before (2009) and after PCV7 introduction (2011) among young children in Cajamarca, Peru, and little change in the overall diversity of circulating serotypes and the number of distinct serotypes detected per child. As expected, carriage rates of vaccine types were reduced, and rates of nonvaccine types increased in 2011 as compared with 2009. The serotypes carried persistently and in high densities in the nasopharynx also shifted, as evidenced by changes in the serotypes classified as persistent and recolonizing in each year. We did not find substantial differences in the densities of serotypes within each cohort; however, the densities of all common serotypes were higher in 2011.
We measured carriage rates of 9.03 and 9.04 detections/person-year in 2009 and 2011, respectively, with serotype-specific carriage rates ranging from 0.0146 to 0.0803 detections/person-month. Previous studies have estimated rates of pneumococcal acquisition, or the rate of colonization in previously uncolonized children. Although the 2 are not directly comparable, as we expect the acquisition rate to be lower than the carriage rate, the comparisons are nonetheless informative. Studies in Kilifi District, Kenya, prior to PCV introduction estimated an acquisition rate of 0.019 acquisitions/person-day among children younger than age 1 year [25], and 0.03 acquisitions/person-day among 3–6-year-olds [26]. These rates are similar to our carriage rate estimate (the 2009 carriage rate we report is equivalent to 0.025 carriage events/person-day). Studies assessing overall carriage rather than new acquisitions have reported lower carriage rates than those estimated from our cohort. A study of Brazilian children under 5 years old reported a 4-month risk of carriage of 74% prior to PCV introduction [27], equivalent to a yearly rate of 4.09 detections [28] and substantially lower than our estimated rates for both before and after PCV introduction. Similarly, studies from Bangladesh [29] and Thailand-Myanmar [11] of PCV-naïve populations report a 2-month colonization risk of approximately 50% in newborns, also equivalent to a yearly rate of slightly more than 4 detections. As noted earlier, there is a wide array of host and environmental factors that influence colonization rates, and we have calculated only unadjusted estimates of carriage in this analysis. However, it is notable that our observed carriage rates in Peru are almost 3 times higher than have been reported in other settings. Reasons for the high burden of Spn carriage in this population of infants and young children in the Peruvian Andes are unclear; however, high altitude and frequent use of indoor wood cook stoves have been shown to increase incidence of viral respiratory infections [30, 31] and may also represent a risk factor for carriage of bacterial organisms. Unfortunately, robust surveillance systems for pneumococcal disease are not in place, so it is difficult to determine whether high carriage in this region may give rise to correspondingly high rates of pneumococcal disease.
We found that over half of all children in both years had the same serotype at first and last detection. 23F, a vaccine serotype, was the only type identified as persistent in 2009, and 6A, 15B, and 19A were frequent recolonizing serotypes in that year. In 2011, 6B and 7C were persistent, and 13 was the only common recolonizer. The observation that persistently colonizing and recolonizing strains in 2011 were unique from those in 2009 suggests not only replacement of vaccine types with nonvaccine types, but possibly a shift in serotype composition and dynamics that reflects a newly developed ability of nonvaccine serotypes to be carried in a more persistent manner.
We did not find substantial changes in density between serotypes within each year, but generally, colonization density in 2011 was higher for all serotypes relative to 2009. In a previous study, with a subset of specimens from the same cohort, we demonstrated that children with higher colonization density were more likely to carry non-PCV7 types, suggesting that the increased circulation of non-PCV7 types in 2011 is driving increases in density [15]. High pneumococcal densities have been shown to be closely correlated with incidence of acute respiratory illness and may facilitate transmission of pneumococci among hosts [32]; however, the present analysis focused on carriage among healthy children. Previous studies have shown in a cohort of children from Portugal the existence of a quantitative “hierarchy” of serotype densities [33], wherein the relative population-level prevalence of serotypes tracks closely with their density in the nasopharynx. We did not observe a clear hierarchy in our cohort in Peru; however, our density measures have some limitations (see below). Moreover, pneumococcal densities are highly dynamic and driven by complex virus-bacteria-host interactions [34, 35], which makes it difficult to draw definitive conclusions about the drivers of changes in bacterial density.
Although densities were reported by serotype, bacterial density was calculated using a quantitative PCR-based method that assayed for a gene (lytA) present in all Spn organisms, and it is therefore not serotype specific. However, a recent study found that in the majority of samples collected from co-colonized individuals, there is one predominant serotype responsible for more than 50% of the bacterial load [21]. Certainly, it is possible that temporal variations in the predominant strain may appear as recolonizing events in our analysis (ie, the last serotype detected is present, but not detected, at first carriage). Although we could not assess co-colonization in our samples, the predominant serotype is likely to be the most epidemiologically relevant, as colonization at higher densities has been associated with the propensity of a serotype to cause disease in the colonized individual [35–39]. Second, our definitions of persistent and recolonizing serotypes are ones that we developed based on only the first and last serotype detected within a child. This fails to capture the small proportion of children who had other serotypes detected in between their first and last samples; however, >80% of children had 2 or fewer distinct serotypes collected. There is some evidence that among healthy children, serotype distributions and density may change with age [8], and we did not account for age differences in our analysis. A subanalysis of children older than 2 years in 2011 showed nearly identical results to that of the full 2011 cohort, so it is unlikely that the addition of new children to the cohort dramatically modified colonization patterns. Last, we are unable to control for secular trends that may have influenced changes in serotype distribution and dynamics either within or across years. Although seasonality may play a role in carriage, we did not have carriage data from the complete year in either 2009 or 2011, and therefore we could not completely assess seasonal trends. Across years, however, there is very low likelihood that any event would alter carriage to a greater extent than PCV7 introduction.
We have shown that overall Spn carriage rates in this population are high and have not appreciably changed after introduction of the PCV7 vaccine; however, the serotypes implicated in persistent and high-density carriage have shifted. Although the transition toward the 13-valent (PCV13) pneumococcal vaccine is currently underway, the epidemiologic effects of PCV7 introduction will continue to be relevant until all countries introduce PCV13; moreover, the results can provide insight into general patterns of carriage changes that might be expected in the context of any pneumococcal vaccine introduction. Further research should examine the effects of PCV13 on complex carriage dynamics and evaluate its impact on the epidemiology of invasive pneumococcal disease.
Supplementary Data
Supplementary materials are available at Open Forum Infectious Diseases online. Consisting of data provided by the authors to benefit the reader, the posted materials are not copyedited and are the sole responsibility of the authors, so questions or comments should be addressed to the corresponding author.
Acknowledgments
We would like to thank Renzo Valeriani, Faidad Khan, and Maneesha Chitanvis from the Rollins School of Public Health at Emory University for their generous assistance in serotyping specimens.
Disclaimer. The content is solely the responsibility of the authors and does not necessarily represent the official view of the National Institutes of Health.
Ethical approval. This study was approved by the Ethical Review Board (ERB) of the Instituto de Investigación Nutricional and the Institutional Review Boards (IRB) of Vanderbilt University and Emory University. An ERB/IRB-approved written informed consent form was obtained from parents of participating subjects at enrollment. The study was also approved by the local health authorities and by community leaders.
Financial support. This work was supported by a Vanderbilt University Clinical and Translational Science Award (National Institutes of Health grant UL1 RR024975 to C.G.G.); the Thrasher Research Fund (grant 02832-9 to C.G.G.); Pfizer investigator-initiated research grants (IIR WS1898786 [0887X1-4492] to C.G.G and IIR WS2079099 to J.E.V.); and the National Institutes of Health (R21AI112768-01A1 to J.E.V.).
Potential conflicts of interest. All authors: no reported conflicts of interest. All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1. Mulholland K. Childhood pneumonia mortality--a permanent global emergency. Lancet 2007; 370:285–9. [DOI] [PubMed] [Google Scholar]
- 2. O’Brien KL, Wolfson LJ, Watt JP et al. ; Hib and Pneumococcal Global Burden of Disease Study Team Burden of disease caused by Streptococcus pneumoniae in children younger than 5 years: global estimates. Lancet 2009; 374:893–902. [DOI] [PubMed] [Google Scholar]
- 3. Bryce J, Boschi-Pinto C, Shibuya K, Black RE; WHO Child Health Epidemiology Reference Group WHO estimates of the causes of death in children. Lancet 2005; 365:1147–52. [DOI] [PubMed] [Google Scholar]
- 4. Wardlaw T, Salama P, Johansson EW, Mason E. Pneumonia: the leading killer of children. Lancet 2006; 368:1048–50. [DOI] [PubMed] [Google Scholar]
- 5. Constenla D, Gomez E, Pio de la Hoz FO et al. The Burden of Pneumococcal Disease and Cost-Effectiveness of a Pneumococcal Vaccine in Latin America and the Caribbean: A Review of the Evidence and a Preliminary Economic Analysis. Washington, DC: The Albert B. Sabin Vaccine Institute Office of International Programs; 2007. [Google Scholar]
- 6. Ochoa TJ, Egoavil M, Castillo ME et al. Invasive pneumococcal diseases among hospitalized children in Lima, Peru. Rev Panam Salud Publica 2010; 28:121–7. [DOI] [PubMed] [Google Scholar]
- 7. Syrjänen RK, Kilpi TM, Kaijalainen TH et al. Nasopharyngeal carriage of Streptococcus pneumoniae in Finnish children younger than 2 years old. J Infect Dis 2001; 184:451–9. [DOI] [PubMed] [Google Scholar]
- 8. Bogaert D, Sluijter M, Toom NL et al. Dynamics of pneumococcal colonization in healthy Dutch children. Microbiology 2006; 152:377–85. [DOI] [PubMed] [Google Scholar]
- 9. Lynch JP, Zhanel GG. Streptococcus pneumoniae: epidemiology and risk factors, evolution of antimicrobial resistance, and impact of vaccines. Curr Opin Pulm Med 2010; 16:217–25. [DOI] [PubMed] [Google Scholar]
- 10. Gray BM, Converse GM, Dillon HC. Epidemiologic studies of Streptococcus pneumoniae in infants: acquisition, carriage, and infection during the first 24 months of life. J Infect Dis 1980; 142:923–33. [DOI] [PubMed] [Google Scholar]
- 11. Turner P, Hinds J, Turner C et al. Improved detection of nasopharyngeal cocolonization by multiple pneumococcal serotypes by use of latex agglutination or molecular serotyping by microarray. J Clin Microbiol 2011; 49:1784–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Leiberman A, Dagan R, Leibovitz E et al. The bacteriology of the nasopharynx in childhood. Int J Pediatr Otorhinolaryngol 1999; 49:S151–3. [DOI] [PubMed] [Google Scholar]
- 13. Weinberger DM, Malley R, Lipsitch M. Serotype replacement in disease after pneumococcal vaccination. Lancet 2011; 378:1962–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Budge PJ, Griffin MR, Edwards KM et al. ; RESPIRA-PERU Group A household-based study of acute viral respiratory illnesses in Andean children. Pediatr Infect Dis J 2014; 33:443–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Hanke CR, Grijalva CG, Chochua S et al. bacterial density, serotype distribution and antibiotic resistance of pneumococcal strains from the nasopharynx of Peruvian children before and after pneumococcal conjugate vaccine 7. Pediatr Infect Dis J 2016; 35:432–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. O’Brien KL, Bronsdon MA, Dagan R et al. Evaluation of a medium (STGG) for transport and optimal recovery of Streptococcus pneumoniae from nasopharyngeal secretions collected during field studies. J Clin Microbiol 2001; 39:1021–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Satzke C, Turner P, Virolainen-Julkunen A et al. ; WHO Pneumococcal Carriage Working Group Standard method for detecting upper respiratory carriage of Streptococcus pneumoniae: updated recommendations from the World Health Organization Pneumococcal Carriage Working Group. Vaccine 2013; 32:165–79. [DOI] [PubMed] [Google Scholar]
- 18. Chien YW, Vidal JE, Grijalva CG et al. Density interactions among Streptococcus pneumoniae, Haemophilus influenzae and Staphylococcus aureus in the nasopharynx of young Peruvian children. Pediatr Infect Dis J 2013; 32:72–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Sakai F, Talekar SJ, Lanata CF et al. ; RESPIRA PERU Group; Investigators Group Expression of Streptococcus pneumoniae virulence-related genes in the nasopharynx of healthy children. PLoS One 2013; 8:e67147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Carvalho Mda G, Tondella ML, McCaustland K et al. Evaluation and improvement of real-time PCR assays targeting lytA, ply, and psaA genes for detection of pneumococcal DNA. J Clin Microbiol 2007; 45:2460–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Sakai F, Chochua S, Satzke C et al. Single-plex quantitative assays for the detection and quantification of most pneumococcal serotypes. PLoS One 2015; 10:e0121064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Sakai F, Sonaty G, Watson D et al. Development and characterization of a synthetic DNA, NUversa, to be used as a standard in quantitative polymerase chain reactions for molecular pneumococcal serotyping. FEMS Microbiol Lett 2017; 364:fnx173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. da Gloria Carvalho M, Pimenta FC, Jackson D et al. Revisiting pneumococcal carriage by use of broth enrichment and PCR techniques for enhanced detection of carriage and serotypes J Clin Microbiol 2010; 48:1611–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Pai R, Gertz RE, Beall B. Sequential multiplex PCR approach for determining capsular serotypes of Streptococcus pneumoniae isolates. J Clin Microbiol 2006; 44:124–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Tigoi CC, Gatakaa H, Karani A et al. Rates of acquisition of pneumococcal colonization and transmission probabilities, by serotype, among newborn infants in Kilifi District, Kenya. Clin Infect Dis 2012; 55:180–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Abdullahi O, Karani A, Tigoi CC et al. Rates of acquisition and clearance of pneumococcal serotypes in the nasopharynges of children in Kilifi District, Kenya. J Infect Dis 2012; 206:1020–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Menezes AP, Azevedo J, Leite MC et al. Nasopharyngeal carriage of Streptococcus pneumoniae among children in an urban setting in Brazil prior to PCV10 introduction. Vaccine 2016; 34:791–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Greenland S. Model-based estimation of relative risks and other epidemiologic measures in studies of common outcomes and in case-control studies. Am J Epidemiol 2004; 160:301–5. [DOI] [PubMed] [Google Scholar]
- 29. Granat SM, Mia Z, Ollgren J et al. Longitudinal study on pneumococcal carriage during the first year of life in Bangladesh. Pediatr Infect Dis J 2007; 26:319–24. [DOI] [PubMed] [Google Scholar]
- 30. Wu A, Budge PJ, Williams J et al. Incidence and risk factors for respiratory syncytial virus and human metapneumovirus infections among children in the remote highlands of Peru. PLoS One 2015; 10:e0130233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Budge PJ, Griffin MR, Edwards KM et al. Impact of home environment interventions on the risk of influenza-associated ARI in Andean children: observations from a prospective household-based cohort study. PLoS One 2014; 9:e91247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Dhoubhadel BG, Yasunami M, Nguyen HAT et al. Bacterial load of pneumococcal serotypes correlates with their prevalence and multiple serotypes is associated with acute respiratory infections among children less than 5 years of age. PLoS One 2014; 9:e110777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Rodrigues F, Danon L, Morales-Aza B et al. Pneumococcal serotypes colonise the nasopharynx in children at different densities. PLoS One 2016; 11:e0163435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Lewnard JA, Givon-Lavi N, Huppert A et al. Epidemiological markers for interactions among Streptococcus pneumoniae, Haemophilus influenzae, and Staphylococcus aureus in upper respiratory tract carriage. J Infect Dis 2016; 213:1596–605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Wolter N, Tempia S, Cohen C et al. High nasopharyngeal pneumococcal density, increased by viral coinfection, is associated with invasive pneumococcal pneumonia. J Infect Dis 2014; 210:1649–57. [DOI] [PubMed] [Google Scholar]
- 36. Chochua S, D'Acremont V, Hanke C et al. Increased nasopharyngeal density and concurrent carriage of Streptococcus pneumoniae, Haemophilus influenzae, and Moraxella catarrhalis are associated with pneumonia in febrile children. PLoS One 2016; 11:e0167725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Vu HTT, Yoshida LM, Suzuki M et al. Association between nasopharyngeal load of Streptococcus pneumoniae, viral coinfection, and radiologically confirmed pneumonia in Vietnamese children. Pediatr Infect Dis J 2011; 30:11–8. [DOI] [PubMed] [Google Scholar]
- 38. Fan RR, Howard LM, Griffin MR et al. Nasopharyngeal pneumococcal density and evolution of acute respiratory illnesses in young children, Peru, 2009–2011. Emerg Infect Dis 2016; 22:1996–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Howard LM, Fan R, Zhu Y et al. Nasopharyngeal pneumococcal density is associated with viral activity but not with use of improved stoves among young Andean children. Open Forum Infect Dis 2017; 4:ofx161. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.