Fig. 7.
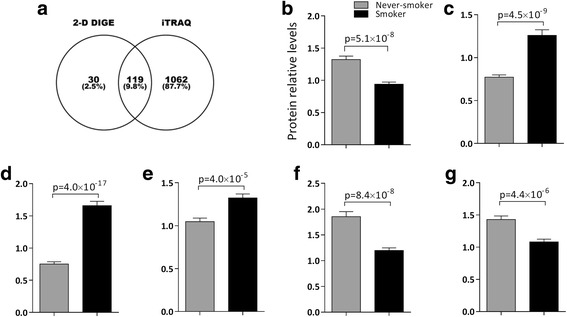
Comparison of the proteomes investigated with the bottom-up iTRAQ peptidomics platform and the top-down 2D–DIGE intact proteomics platform. a) Venn diagram displaying the overlap of proteins identified with the two platforms indicates that the majority of proteins identified with iTRAQ (90%) were novel as compared to 2D–DIGE. The proteins that overlapped were generally altered in a similar fashion, thereby providing validation across platforms. b) Protein 60S acidic ribosomal protein P0 with decreased level in ribosomal pathway in Smokers (2-D DIGE: p = 5.1 × 10−8; iTRAQ: p = 0.009); c) Protein Cytochrome b-c1 complex subunit Rieske with elevated level in pathway oxidative phosphorylation in Smokers (2-D DIGE: p = 4.5 × 10−9; iTRAQ: p = 1.5 × 10−5); d) Protein Cathepsin D with increased level in lysosomal pathway in Smokers (2D DIGE: p = 4.0 × 10−17; iTRAQ: p = 4.3 × 10−19); e) ATP synthase subunit d in mitochondria with upregulated level in citrate cycle in Smokers (2-D DIGE: p = 4.0 × 10−5; iTRAQ: p = 6.3 × 10−5); f) Enzyme Aldehyde dehydrogenase in mitochondria with downregulated level in pathway valine, leucine and isoleucine degradation in Smokers (2-D DIGE: p = 8.4 × 10−8; iTRAQ: p = 0.02); g) Protein actin, cytoplasmic 1 with decreased level in pathway phagosome in smokers (2-D DIGE: p = 4.4 × 10−6; iTRAQ: p = 0.007)
