Fig. 6.
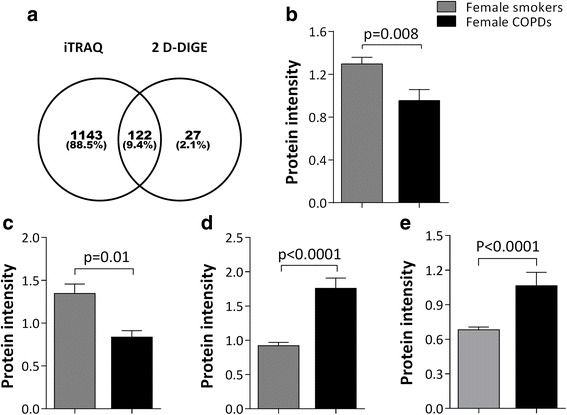
Comparison of results from 2D–DIGE intact proteomics [15] and iTRAQ shotgun proteomics analysis on the same cohort. a) Venn diagram showing that the majority of protein species identified by 2D–DIGE overlapped with proteins identified by iTRAQ. Ninety percent of the proteins identified by iTRAQ were not identified by 2D–DIGE. However, several of the proteins identified by 2D–DIGE served as validation of key proteins in pathways found to be enriched in the current study: Actin-related protein 3 (ARP3; panel b;, 2D–DIGE: p = 0.008; iTRAQ: p = 0.01, is involved in the pathways FcγR-mediated phagocytosis and regulation of actin cytoskeleton; Beta-hexosaminidase subunit beta (HEXB; panel c;)2D–DIGE: p = 0.01; iTRAQ: p = 0.02,); and ATP synthase subunit beta (ATP5B, panel d;) (2D–DIGE: p < 0.0001; p = 0.04,) are both involved in the lysosomal- and oxidative phosphorylation pathways,; e), Leukotriene A-4 hydrolase (LTA4H) (2D–DIGE: p < 0.0001; iTRAQ: p = 0.01,). is one of the most prominent proteins for driving the separation between female COPD and Smokers groups by both proteomics platforms. Data in b, c, d and e are expressed as mean ± SE
