Figure 4. NP-protein interactions by surface plasmon resonance.
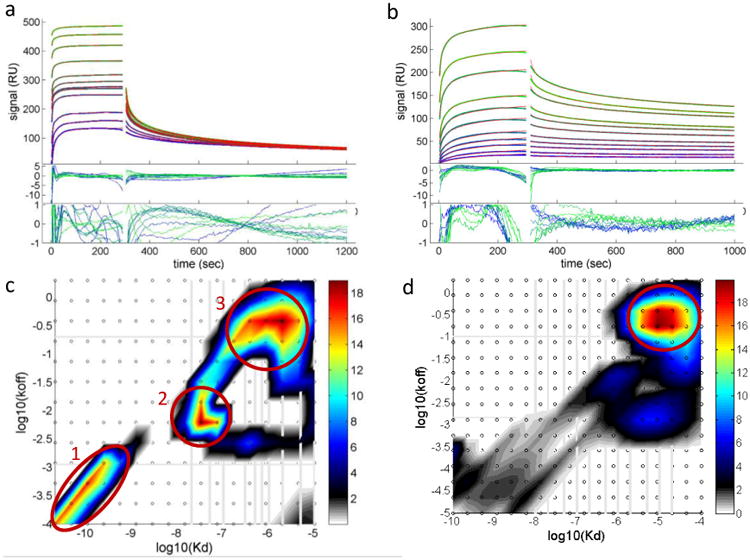
a, Analysis of AuMBA-CrataBL binding traces with the surface-site distribution model. b, The same for AuGSH-CrataBL. Phosphate buffer supplemented with 150 mM NaCl was used as running buffer. AuMBA was injected in the flow at the concentrations of 0.02, 0.05, 0.1, 0.3, 0.5 (2×), 0.7, 1, 2, 5, 10, 20 μM. For AuGSH the concentration range was 0.01, 0.02, 0.05, 0.1, 0.2, 0.5, 1, 2, 5, 10, 20 μM. Shown are the experimental traces (green and blue lines), best-fit curves (red lines), and fitting residuals. c, Calculated affinity and rate constant distributions for AuMBA-CrataBL. d, The same for AuGSH-CrataBL. Circled regions indicate the major peaks in the distributions. Integration of the peaks provides the binding parameters KD, kon and koff.
