Extended Data Figure 6.
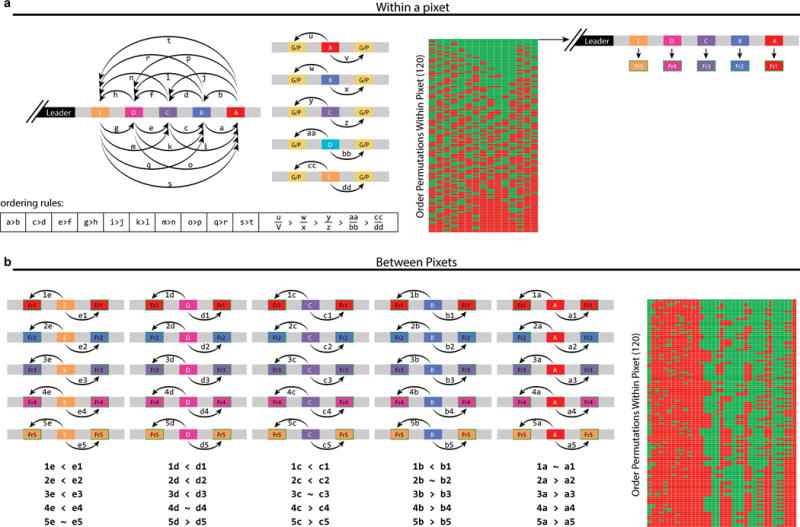
Recall of frame order over time based on position in the CRISPR array. a. Initial set of rules to test the order of spacers within a pixet. Every time two spacers from the same pixet are found in a single array, their relative physical location (with respect to the leader) is extracted. As is the location of each spacer relative to spacers drawn from the genome or plasmid (G/P). The actual sequence of electroporated protospacers should occupy arrays in a predictable physical arrangement, as described by these ordering rules. Every possible permutation of spacers within a pixet is tested against each of these rules and, if a permutation satisfies all the rules, spacers are assigned to frame. b. Second set of tests to compare between pixets. If no permutation satisfies all of the tests in a, spacers are compared to previously assigned spacers from other pixets pairwise when found in the same array. A larger set of rules will hold true for the actual sequence of electroporated protospacers when compared against previously assigned spacers. Again, all possible order permutations are tested, and order is assigned based on the best overall satisfaction of these ordering rules.
