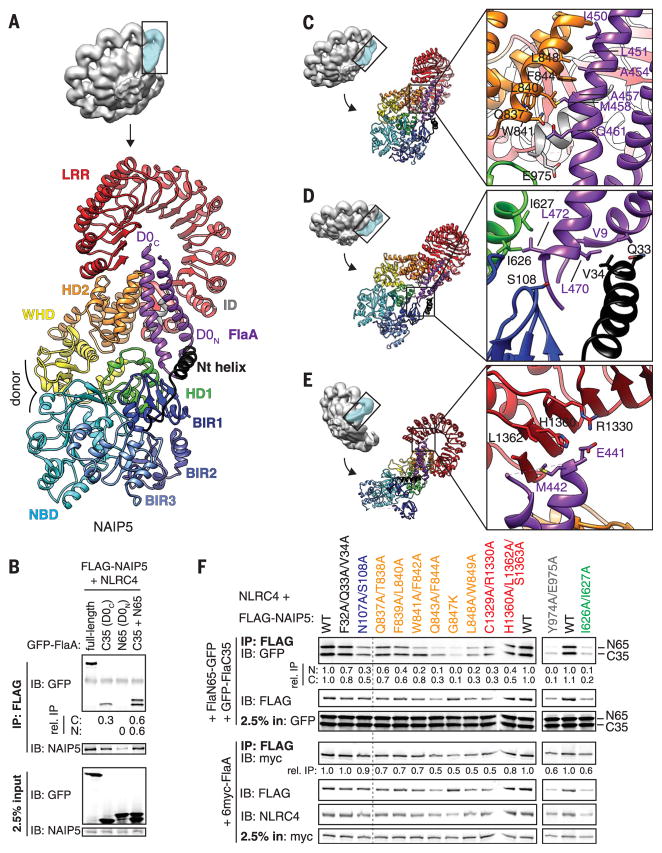
Fig. 2. Multiple NAIP5 domains contact extended surfaces on both helices of the flagellin D0 domain.
(A) The D0 domain (purple) is locked into place by multiple NAIP5 domains. (B) Both D0 helices bind to NAIP5. (C to E) Detailed interactions between the flagellin D0 helices and the NAIP5 domains HD2 and ID (C); Nt, BIR1, and HD1 (D); and LRR (E). Side chains shown correspond to mutated residues in (F). (F) Mutagenesis confirms the importance of NAIP5 residues in binding D0N, D0C, or full-length FlaA. Point mutations in NAIP5 residues that contact D0C disrupt both D0N and D0C binding because association of D0N requires D0C binding [(B) and fig. S9A]. In (B) and (F), relative IP strength was quantified by densitometry [IP signal normalized to input and then GFP-FlaA (B) or WT NAIP5 (F)]. Results are representative of at least three independent experiments. IP, immunoprecipitation; IB, immunoblot; WT, wild type. Single-letter abbreviations for the amino acid residues are as follows: A, Ala; C, Cys; D, Asp; E, Glu; F, Phe; G, Gly; H, His; I, Ile; K, Lys; L, Leu; M, Met; N, Asn; P, Pro; Q, Gln; R, Arg; S, Ser; T, Thr; V, Val; W, Trp; and Y, Tyr.