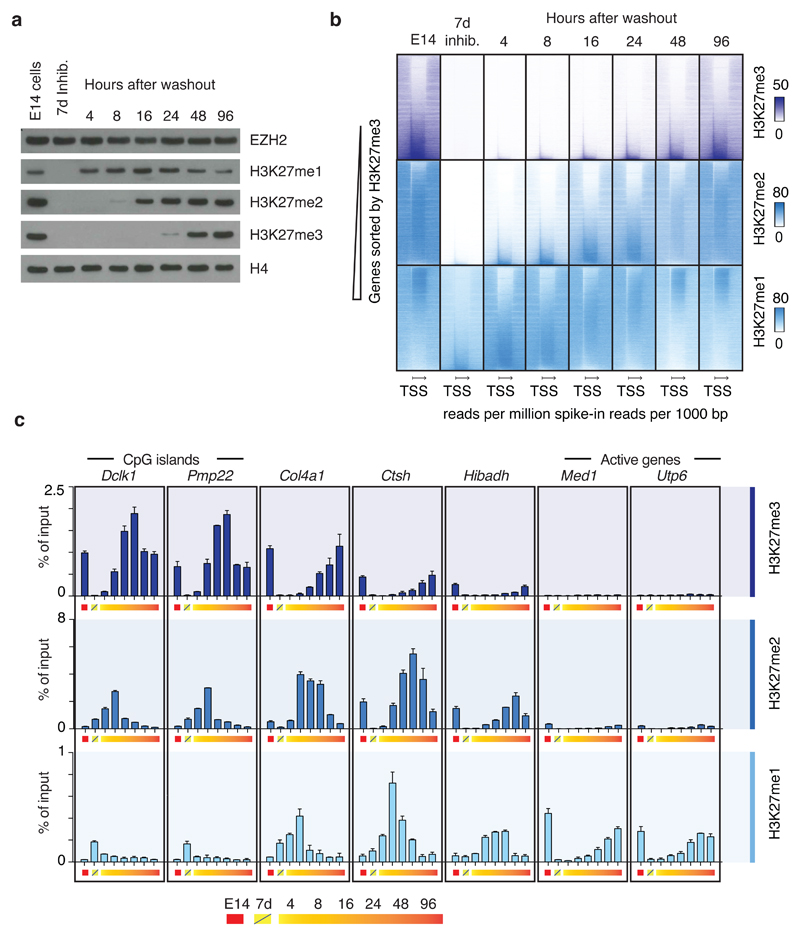
Figure 3. Regional de novo methylation kinetics.
a, Western blots of extracts from E14 cells treated with 10 µM EPZ6438 EZH2 inhibitor for 7 days and sampled at time points following inhibitor washout. Blots were probed with the indicated antibodies. Experiment has been repeated at least three times with same result. Uncropped Western blot images are shown in Supplementary Data Set 1
b, Heatmaps of spike-in normalized ChIP-seq data probing restoration kinetics of H3K27me3 (top row), H3K27me2 (middle row) and H3K27me1 (bottom row) in genic regions.
c, ChIP-qPCR data probing kinetics of H3K27me3 (top row), H3K27me2 (middle row) and H3K27me1 (bottom row) at seven representative loci labeled (above graph) according to nearest gene and in approximate order of decreasing methylation rates (% of input/hr washout). Data values come from a single biological (n=1) experiment, with error bars depicting s.d. of technical duplicates. Data from a biological replicate of the experiment is shown in Supplementary Fig. 4c, and the experiments have also been performed once in Ezh2f/f and Ezh1 KO mESC lines with comparable result.