Abstract
The aryl hydrocarbon receptor (AHR) is an important component of the host-microbiota communication network. Comparisons of wild-type and Ahr-null mice as well as from exposure studies with potent AHR ligands (e.g., 2,3,7,8-tetrachlorodibenzo-p-dioxin) have provided compelling evidence that the AHR may be a master regulator of the host-microbiota interaction thus helping to shape the immune system and impact host metabolism. Metabolomics and sequenced-based microbial community profiling, two recent technological advances, have helped to solidify this host-microbiota signaling concept and identified not only how specific ligands generated by the host and by the microbiota can activate the AHR, but also how activation/disruption of the AHR can influence and shape the microbiota. We are just beginning to understand how the temporal nature and tissue- and microbiota-specific generation of AHR ligands contribute to many AHR-dependent processes. In this review, we focus on several recent advances where metabolomics and characterization of the microbiota structure and function have generated new perspectives by which to evaluate AHR activity.
Keywords: AHR, microbiota, microbiome, metabolomics, mass spectrometry
INTRODUCTION
The aryl hydrocarbon receptor (AHR) is a key regulator of the response to drugs and toxicants, is important in immune system development and homeostasis (reviewed in [1]), and, more recently, AHR activity has been reported to modulate the microbiota residing on the skin [2] and in the gut [3–5]. The diverse roles of the AHR are driven in large part by a similarly diverse set of ligands (reviewed in [6]) including xenobiotics (e.g., environmental contaminants such as 2,3,7,8-tetrachlorodibenzo-p-dioxin [TCDD], benzo[a]pyrene), diet-derived chemicals (e.g., flavonoids and indoles), endogenous (e.g., 6-formylindolo[3,2-b]carbazole [FICZ]), and bacterial-associated or -produced metabolites (e.g., phenazines, tryptophan catabolites). Within the last ten years we have witnessed a renewed interest in the AHR moving beyond its well-studied role as a xenobiotic sensor to new roles that have implications in host metabolism, barrier organ function (reviewed in [7]), and, importantly, to roles that may represent new therapeutic targets for several human diseases including cancer and obesity [8].
Understanding the AHR has been advanced through the development of tools including mouse models, highly sensitive reporter cell lines, and through the generation and characterization of diverse AHR ligands with differential affinity. In particular, the strategic use of tissue-specific knockout mice [9] (generated in the low affinity Ahrd background) has clarified the role of AHR in different tissue compartments including immune cell populations, hepatocytes, keratinocytes, and the intestinal epithelium. However, differences in ligand binding affinity between the various Ahr alleles in mice (Ahrd vs Ahrb) and important species differences between human, mouse, and other model organisms have yet to be fully reconciled in the literature [10] and (reviewed in [11]). Sensitive reporter lines have helped not only to identify new AHR ligands but have also been instrumental for monitoring low-level exposure to toxic environmental AHR ligands [12]. Despite these incredibly valuable tools, our knowledge of the quantity, identity, and ultimate distribution of AHR endogenous as well as exogenous ligands throughout the body remains limited.
With cutting-edge tools including metabolomics (i.e., chemical fingerprinting) and sequenced-based microbial community profiling, new and exciting roles for the AHR are beginning to unfold. In this review we highlight and discuss how metabolomics and characterization of the microbiota has helped to advance understanding of AHR activity and function, and provide our vision for ways to clarify the role of AHR in modulating the host-microbiota relationship.
AHR and METABOLOMICS
Metabolomics has accelerated numerous discoveries in the fields of toxicology, drug metabolism, and receptor biology [13–15]. For example, metabolomics has been instrumental in uncovering the important contribution of the gut microbiota to host metabolism [16] and has provided detailed metabolic maps of AHR ligands [17]. Metabolomics is typically conducted by hyphenated techniques such as liquid or gas chromatography coupled with mass spectrometry and by nuclear magnetic resonance spectroscopy (NMR). It is important to emphasize that given the varied physicochemical properties of chemicals making up the metabolome numerous platforms are required, which is especially true when considering the diverse array of known AHR ligands (reviewed in [18]). Below we critically evaluate the contributions that metabolomics has made to the AHR field by examining reports from mouse models and human studies.
Mouse Models
Metabolomic studies are profoundly impacted by age, sex, diet, lifestyle, and environmental exposures (reviewed in [13]). Therefore, mouse models are essential to study the metabolic impact of AHR activation without the influence of confounding factors. Many of the early metabolomic studies with AHR surround the prototypical AHR ligand TCDD at generally high doses. For example, Matsubara and colleagues [19] examined the dose- and time-dependent impact of TCDD on the mouse serum metabolome. The authors gave TCDD to C57Bl/6N mice via intraperitoneal injection (10 μg/kg and 200 μg/kg) and using ultra-high pressure liquid chromatography coupled with quadrupole time-of-flight mass spectrometry (UHPLC-QTOFMS) were able to identify specific upregulation of azaleic acid mono-esters, which was attributed to downregulation of hepatic carboxylesterase 3 (CES3) activity. It was concluded that downregulation of CES3 was associated with steatohepatitis which is commonly observed with high doses of TCDD and related environmental contaminants. Other examples using high dose TCDD include comparison of C57Bl/6 and DBA/2 mice (representing the high affinity and low affinity alleles, respectively) exposed to TCDD (20 μg/kg p.o. daily for 7 days) [20]. The authors identified using QTOFMS-based metabolomics significant accumulation of liver fatty acids and lysophosphocholines in the high affinity C57Bl/6 mice. The DBA/2 mice exhibited a significant, but blunted response to TCDD exposure. Similarly, a series of studies elegantly examined the longitudinal metabolic perturbations associated with TCDD-associated fibrosis in female C57Bl/6 mice using a variety of high throughput techniques including microarray, RNA-seq, and metabolomics [21–23]. Mice were given TCDD via gavage at doses ranging from 0.01 μg/kg to 30 μg/kg every four days over a period of 28 or 92 days. Using a targeted metabolite profiling approach, the authors report that TCDD exposure significantly perturbed many metabolic pathways including glycogen, amino acid, TCA, and lipid metabolism; however, significant changes were only observed at doses >1 μg/kg. In addition, 1H NMR-based metabolomic studies of male wild-type and Ahr−/− C57Bl/6J mice treated with 24 μg/kg 2,3,7,8-tetrachlorodibenzofuran (TCDF), another typical environmental AHR ligand, demonstrated that TCDF exposure profoundly affects host metabolic pathways such as hepatic lipogenesis, glucose and energy metabolism, and de novo fatty acid biosynthesis [3, 24].
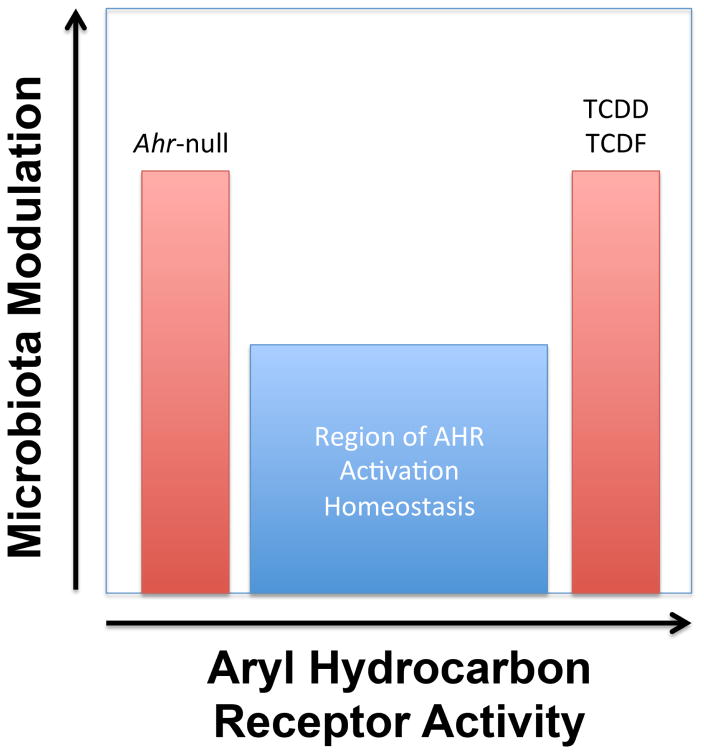
Collectively, these as well as other investigations of AHR activation point to a central role for AHR in hepatic lipid metabolism [25, 26]. However, interpreting these data in contexts outside of industrial accidents or intentional poisonings remains challenging given that TCDD burden can be orders of magnitude greater than what might be found in the general population. Therefore, studies with TCDD as well as other relevant environmental AHR ligands including polychlorinated biphenyls and polycyclic aromatic hydrocarbons should be investigated as doses resembling those found in the general population to best appreciate their metabolic impact. Longitudinal dose-response studies comparing the most toxic AHR ligands (e.g., TCDD) with those generally considered potentially important for health (e.g., indole-3-carbinol, a breakdown product of the glucosinolate glucobrassicin found in cruciferous vegetables) would be invaluable to differentiate the metabolic effects of AHR-mediated toxicity from AHR health promoting effects (Figure 1).
Figure 1.
Studies have demonstrated that both genotype and potent activation of the AHR can lead to pronounced changes in the microbiota community structure and function. Evidence from metabolomics suggests that outside the region of AHR activation homeostasis, metabolic abnormalities are likely to be observed. For example, activation of AHR with the potent agonists TCDD or TCDF promote increases in hepatic lipogenesis, alterations in bile acid pools, and increased production of short chain fatty acids by the gut microbiota. The challenge presented to the field is defining what level of AHR activation and by which ligands are important for promoting health.
Human Studies
To date, there have been only a few studies that have attempted to identify how exposure to environmental AHR ligands influences the metabolome. For example, the serum metabolome from 81 Dutch chlorophenoxy herbicide factory workers and 63 non-exposed workers was measured via UHPLC-QTOFMS [27]. While significant differences in blood TCDD concentrations were observed (2.09 parts per trillion [ppt] compared with 0.44 ppt), the authors reported that no metabolic features (here a metabolic feature is defined as a mass and retention pair that has not been structurally elucidated) survived false discovery rate correction. Limitations of the study included its retrospective nature (workers were exposed from 1953–1969 but blood was sampled in 2007–2008), differences in age between the exposed and non-exposed cohorts, and the rather limited metabolomic analysis (the samples were only analyzed by one platform in a single ionization mode). A retrospective urinary metabolomic investigation was conducted in Czech factory workers producing the herbicide trichlorophenol acetic acid [28]. Using UHPLC-QTOFMS-based metabolomics, the authors report significant alterations in steroid and bile acid metabolism and interestingly were able to find a subset of these metabolites were enriched in urine samples obtained from Victor Yushchenko who was poisoned with TCDD. However, it is important to note several limitations associated with this study including its rather small sample size (only 11 workers were studied nearly 40 years after the exposure) but recent examination in an independent cohort from workers exposed via municipal waste incineration suggest that these metabolic perturbations are related to TCDD exposure [29]. Based on these studies it is tempting to speculate that TCDD and other potent environmental AHR ligands may impart a metabolic fingerprint that could be important for elucidating the mechanisms behind many of the metabolic diseases thought to be associated with environmental pollutants. Whether or not certain AHR ligands are causal agents in metabolic disease, especially as it relates to perturbation of the microbiome, remains an underexplored area of investigation. However, as is discussed below, emerging evidence from animal models is beginning to support the concept that the microbiome and the AHR may work in concert to promote human metabolic disease.
AHR and the MICROBIOME
In 1991, Perdew and Babbs reported that rat fecal suspensions incubated with tryptophan or indole-3-carbinol could generate AHR ligands thus providing some of the first evidence that AHR activity in the gastrointestinal tract could be modulated by bacterial metabolism [30]. Interestingly, several metabolomics investigations hinted at a potential connection between AHR and the gut microbiota. Tryptophan catabolites such as indole-3-aldehyde, an AHR ligand, were found to be produced by Lactobacillus reuteri and, importantly, modulated the AHR-Il-22 signaling axis [5]. Others have similarly reported that tryptophan catabolites (reviewed in [31]) have important antimicrobial, immunoregulatory, and disease-resistance effects that all appear to be mediated, at least in part, by the AHR.
16S rRNA gene sequencing has afforded the opportunity to perform census taking on the bacteria population residing in or on the organism. With respect to the AHR, several recent studies have identified that not only does AHR activation with potent agonists like TCDF [3], TCDD [4], or tryptophan catabolites [32] promote changes in the gut microbiota population, but also the AHR genotype can influence the bacterial population residing in the gut [3, 11, 33] and also on the skin [2]. Studies with environmental contaminants like TCDD or TCDF suggest that these compounds can influence the gut microbiota population in an Ahr-dependent manner. However, like the metabolomic studies described above, the changes in the gut microbiota structure and function have been reported after mice received relatively high doses via the diet. Combining 16S rRNA gene sequencing analysis with metabolomics has been critically important to understand the functional implications of changing the gut microbiota population. For example, mice receiving TCDF (24 μg/kg p.o. daily for 5 days) demonstrated a pronounced shift in the cecal gut microbiota community structure as well as having decreased levels of ileum segmented filamentous bacteria. Metabolomics analysis of cecal extracts identified increased production of short chain fatty acids that are known to be important for activation of hepatic de novo lipogenesis [3]. Taken together with other metabolomic investigations, it appears that at least part of the AHR-associated increases in hepatic lipogenesis can be attributed to the gut microbiota. Clearly, additional investigation into the timing, dose, route of exposure, and nature of the AHR ligand (health promoting or toxicant) must be conducted in order to better understand how the AHR and the microbiota may contribute to health and disease.
Importantly, development of assays that specifically address microbiome toxicity will be an important component to understand how AHR ligands (and other drugs or toxicants) directly affect the microbiome. For example, sophisticated studies [34, 35] using flow cytometry-based assays were developed to quantify the metabolic activity and cell damage of gut microbes after exposure to known microbial poisons like antibiotics but also to host target drugs (e.g., digoxin). Further refinement of assays to assess microbiome toxicity will be key to advancing our understanding for how AHR ligands might directly influence the microbiota.
FUTURE STUDIES TO ADVANCE OUR UNDERSTANDING OF AHR ACTIVITY AND FUNCTION
While there are extensive datasets generated from gene expression studies (e.g., microarray and RNA-seq), no studies to date have attempted to compare the metabolic signatures associated with AHR activation by diverse agonists (Figure 2). Moreover, many of these studies have focused solely on high, sometimes physiologically improbable, doses. While there are examples comparing potent agonists like FICZ and TCDD in systems like the chicken embryo [36] or under conditions associated with influenza infection [37], carefully controlled studies that incorporate metabolomics and 16S rRNA gene sequencing have yet to be accomplished. Why is this important? For example, might we expect potent agonists like TCDD or FICZ to exhibit the same metabolomic profile or can distinct metabolic profiles be obtained that differentiate toxicity from health promoting benefits? Suggestions from studies in Hepa1c1c7 and C57Bl/6 mice exposed to TCDD, 3,3′,4,4′,5-pentachlorobiphenyl (PCB-126), β-naphthoflavone, and indolo[3,2-b]carbazole suggest that indeed selective AHR modulation might exert different metabolic effects [38]. These studies of course are not without their complications given that dose considerations must take into account factors like differential binding affinity as well as half-life. Further while tryptophan catabolites are not only generated by the gut microbiota they also appear to influence the gut microbiota population. Therefore, how might other diet-derived chemicals like indole-3-carbinol influence the gut microbiota structure and function? While it is clear that acute doses of AHR agonists can profoundly influence host and gut microbiota metabolism, it remains to be determined how chronic, low dose exposure influences these organ systems [39]. Importantly, we do not fully appreciate how timing of these exposures (both from environmental as well as from the diet) may influence the gut microbiota. Given what we know regarding early life antibiotic exposure [40], might the timing of AHR activation in the gut or the skin or lung influence the microbiota as it establishes itself early in the life of the host? Interestingly, Lithobates pipiens larval exposure to PCB-126 resulted in significant and permanent changes in the gut microbiota of adult frogs [41]. However, it is not clear if these effects were mediated by direct impact on the microbiota or driven by host physiology. These observations not only underscore the potential for these compounds to disrupt early life colonization but also suggest that in addition to human exposure, the impact of these environmental chemicals on the microbiome of diverse species should also be investigated.
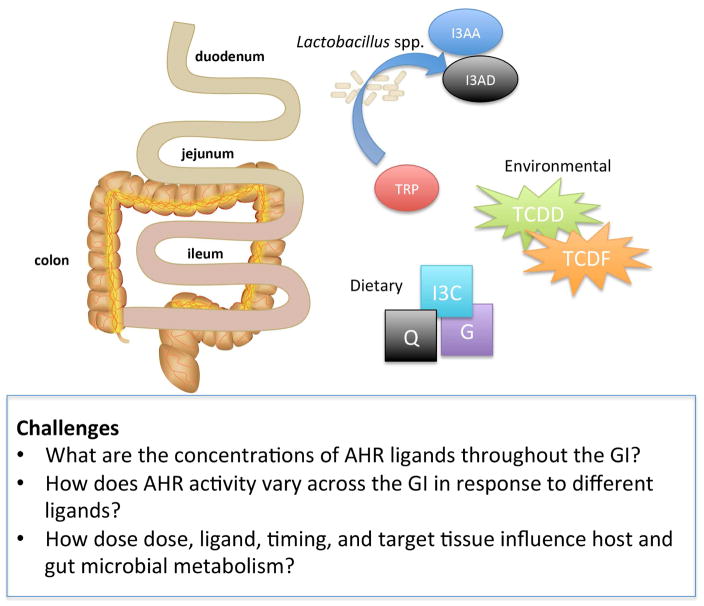
Figure 2.
AHR ligands include those produced by microbiota metabolism of tryptophan (TRP, tryptophan; I3AD, indole-3-aldehyde; I3AA, indole-3-acetic acid), those obtained through the diet (I3C, indole-3-carbinol; Q, quercetin; G, galangin), or those from environmental exposure (TCDD, TCDF). With respect to the gastrointestinal tract, challenges are proposed to further clarify how the location and concentration of endogenous and xenobiotic AHR ligands influence gut metabolism including that of the gut microbiota.
In summary, it is clear that technology has driven many discoveries in the AHR field. The combined application of multiple ‘omics’ approaches including genomics, transcriptomics, proteomics, metabolomics, and using approaches to catalog the microbiota will be important to further elucidate AHR-dependent mechanisms as well as to provide new perspectives by which to understand toxicity. It will only be through careful study design and the strategic use of both genetic and pharmacologic (e.g., agonists, antagonists, or selective AHR modulators) manipulation of AHR activity will we gain a deeper understanding for how the AHR and the microbiota interact to promote health and disease.
Highlights.
Metabolomic approaches have clarified the metabolic consequences of AHR activation.
AHR genotype and activation by diverse ligands impacts the microbiota.
Timing, dose, exposure route, and type of AHR ligand are areas to further explore.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Stockinger B, Di Meglio P, Gialitakis M, Duarte JH. The aryl hydrocarbon receptor: multitasking in the immune system. Annu Rev Immunol. 2014;32:403–32. doi: 10.1146/annurev-immunol-032713-120245. [DOI] [PubMed] [Google Scholar]
- 2.Haas K, Weighardt H, Deenen R, Kohrer K, Clausen B, Zahner S, Boukamp P, Bloch W, Krutmann J, Esser C. Aryl Hydrocarbon Receptor in Keratinocytes Is Essential for Murine Skin Barrier Integrity. J Invest Dermatol. 2016 doi: 10.1016/j.jid.2016.06.627. [DOI] [PubMed] [Google Scholar]
- •3.Zhang L, Nichols RG, Correll J, Murray IA, Tanaka N, Smith PB, Hubbard TD, Sebastian A, Albert I, Hatzakis E, Gonzalez FJ, Perdew GH, Patterson AD. Persistent Organic Pollutants Modify Gut Microbiota-Host Metabolic Homeostasis in Mice Through Aryl Hydrocarbon Receptor Activation. Environ Health Perspect. 2015;123(7):679–88. doi: 10.1289/ehp.1409055. Identified that the environmental AHR ligand TCDF impacts the microbiome in an Ahr-dependent manner. Used metabolomics analysis of cecal extracts to evaluate the metabolic significant of these gut microbial changes. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lefever DE, Xu J, Chen Y, Huang G, Tamas N, Guo TL. TCDD modulation of gut microbiome correlated with liver and immune toxicity in streptozotocin (STZ)-induced hyperglycemic mice. Toxicol Appl Pharmacol. 2016;304:48–58. doi: 10.1016/j.taap.2016.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zelante T, Iannitti RG, Cunha C, De Luca A, Giovannini G, Pieraccini G, Zecchi R, D’Angelo C, Massi-Benedetti C, Fallarino F, Carvalho A, Puccetti P, Romani L. Tryptophan catabolites from microbiota engage aryl hydrocarbon receptor and balance mucosal reactivity via interleukin-22. Immunity. 2013;39(2):372–85. doi: 10.1016/j.immuni.2013.08.003. [DOI] [PubMed] [Google Scholar]
- 6.Murray IA, Patterson AD, Perdew GH. Aryl hydrocarbon receptor ligands in cancer: friend and foe. Nat Rev Cancer. 2014;14(12):801–14. doi: 10.1038/nrc3846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Esser C, Rannug A. The aryl hydrocarbon receptor in barrier organ physiology, immunology, and toxicology. Pharmacol Rev. 2015;67(2):259–79. doi: 10.1124/pr.114.009001. [DOI] [PubMed] [Google Scholar]
- 8.Moyer BJ, Rojas IY, Kerley-Hamilton JS, Hazlett HF, Nemani KV, Trask HW, West RJ, Lupien LE, Collins AJ, Ringelberg CS, Gimi B, Kinlaw WB, 3rd, Tomlinson CR. Inhibition of the aryl hydrocarbon receptor prevents Western diet-induced obesity. Model for AHR activation by kynurenine via oxidized-LDL, TLR2/4, TGFbeta, and IDO1. Toxicol Appl Pharmacol. 2016;300:13–24. doi: 10.1016/j.taap.2016.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Walisser JA, Glover E, Pande K, Liss AL, Bradfield CA. Aryl hydrocarbon receptor-dependent liver development and hepatotoxicity are mediated by different cell types. Proc Natl Acad Sci U S A. 2005;102(49):17858–63. doi: 10.1073/pnas.0504757102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Flaveny CA, Murray IA, Chiaro CR, Perdew GH. Ligand selectivity and gene regulation by the human aryl hydrocarbon receptor in transgenic mice. Mol Pharmacol. 2009;75(6):1412–20. doi: 10.1124/mol.109.054825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hubbard TD, Murray IA, Perdew GH. Indole and Tryptophan Metabolism: Endogenous and Dietary Routes to Ah Receptor Activation. Drug Metab Dispos. 2015;43(10):1522–35. doi: 10.1124/dmd.115.064246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Brennan JC, He G, Tsutsumi T, Zhao J, Wirth E, Fulton MH, Denison MS. Development of Species-Specific Ah Receptor-Responsive Third Generation CALUX Cell Lines with Enhanced Responsiveness and Improved Detection Limits. Environ Sci Technol. 2015;49(19):11903–12. doi: 10.1021/acs.est.5b02906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Johnson CH, Patterson AD, Idle JR, Gonzalez FJ. Xenobiotic metabolomics: major impact on the metabolome. Annu Rev Pharmacol Toxicol. 2012;52:37–56. doi: 10.1146/annurev-pharmtox-010611-134748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Patterson AD, Gonzalez FJ, Idle JR. Xenobiotic metabolism: a view through the metabolometer. Chem Res Toxicol. 2010;23(5):851–60. doi: 10.1021/tx100020p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Montanez JE, Peters JM, Correll JB, Gonzalez FJ, Patterson AD. Metabolomics: an essential tool to understand the function of peroxisome proliferator-activated receptor alpha. Toxicol Pathol. 2013;41(2):410–8. doi: 10.1177/0192623312466960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- •16.Wikoff WR, Anfora AT, Liu J, Schultz PG, Lesley SA, Peters EC, Siuzdak G. Metabolomics analysis reveals large effects of gut microflora on mammalian blood metabolites. Proc Natl Acad Sci U S A. 2009;106(10):3698–703. doi: 10.1073/pnas.0812874106. Demonstrated that the gut microbiota significantly contribute to host metabolism. Serves as one of the primary examples where metabolomics was useful in identifying microbial metabolites. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fang ZZ, Krausz KW, Nagaoka K, Tanaka N, Gowda K, Amin SG, Perdew GH, Gonzalez FJ. In vivo effects of the pure aryl hydrocarbon receptor antagonist GNF-351 after oral administration are limited to the gastrointestinal tract. Br J Pharmacol. 2014;171(7):1735–46. doi: 10.1111/bph.12576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Denison MS, Nagy SR. Activation of the aryl hydrocarbon receptor by structurally diverse exogenous and endogenous chemicals. Annu Rev Pharmacol Toxicol. 2003;43:309–34. doi: 10.1146/annurev.pharmtox.43.100901.135828. [DOI] [PubMed] [Google Scholar]
- 19.Matsubara T, Tanaka N, Krausz KW, Manna SK, Kang DW, Anderson ER, Luecke H, Patterson AD, Shah YM, Gonzalez FJ. Metabolomics identifies an inflammatory cascade involved in dioxin- and diet-induced steatohepatitis. Cell Metab. 2012;16(5):634–44. doi: 10.1016/j.cmet.2012.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu S, Lu F, Wang X, Sun W, Chen P, Dong W. Metabolomic study of a rat fever model induced with 2,4-dinitrophenol and the therapeutic effects of a crude drug derived from Coptis chinensis. Am J Chin Med. 2011;39(1):95–109. doi: 10.1142/S0192415X11008671. [DOI] [PubMed] [Google Scholar]
- 21.Fader KA, Nault R, Ammendolia DA, Harkema JR, Williams KJ, Crawford RB, Kaminski NE, Potter D, Sharratt B, Zacharewski TR. 2,3,7,8-Tetrachlorodibenzo-p-Dioxin Alters Lipid Metabolism and Depletes Immune Cell Populations in the Jejunum of C57BL/6 Mice. Toxicol Sci. 2015;148(2):567–80. doi: 10.1093/toxsci/kfv206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nault R, Fader KA, Ammendolia DA, Dornbos P, Potter D, Sharratt B, Kumagai K, Harkema JR, Lunt SY, Matthews J, Zacharewski T. Dose-dependent metabolic reprogramming and differential gene expression in TCDD-elicited hepatic fibrosis. Toxicol Sci. 2016 doi: 10.1093/toxsci/kfw163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nault R, Fader KA, Kirby MP, Ahmed S, Matthews J, Jones AD, Lunt SY, Zacharewski TR. Pyruvate Kinase Isoform Switching and Hepatic Metabolic Reprogramming by the Environmental Contaminant 2,3,7,8-Tetrachlorodibenzo-p-Dioxin. Toxicol Sci. 2016;149(2):358–71. doi: 10.1093/toxsci/kfv245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang L, Hatzakis E, Nichols RG, Hao R, Correll J, Smith PB, Chiaro CR, Perdew GH, Patterson AD. Metabolomics Reveals that Aryl Hydrocarbon Receptor Activation by Environmental Chemicals Induces Systemic Metabolic Dysfunction in Mice. Environ Sci Technol. 2015;49(13):8067–77. doi: 10.1021/acs.est.5b01389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lee JH, Wada T, Febbraio M, He J, Matsubara T, Lee MJ, Gonzalez FJ, Xie W. A novel role for the dioxin receptor in fatty acid metabolism and hepatic steatosis. Gastroenterology. 2010;139(2):653–63. doi: 10.1053/j.gastro.2010.03.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tanos R, Murray IA, Smith PB, Patterson A, Perdew GH. Role of the Ah receptor in homeostatic control of fatty acid synthesis in the liver. Toxicol Sci. 2012;129(2):372–9. doi: 10.1093/toxsci/kfs204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hosnijeh FS, Pechlivanis A, Keun HC, Portengen L, Bueno-de-Mesquita HB, Heederik D, Vermeulen R. Serum metabolomic pertubations among workers exposed to 2,3,7,8-tetrachlorodibenzo-p-dioxin (TCDD) Environ Mol Mutagen. 2013;54(7):558–565. doi: 10.1002/em.21802. [DOI] [PubMed] [Google Scholar]
- 28.Jeanneret F, Boccard J, Badoud F, Sorg O, Tonoli D, Pelclova D, Vlckova S, Rutledge DN, Samer CF, Hochstrasser D, Saurat JH, Rudaz S. Human urinary biomarkers of dioxin exposure: analysis by metabolomics and biologically driven data dimensionality reduction. Toxicol Lett. 2014;230(2):234–43. doi: 10.1016/j.toxlet.2013.10.031. [DOI] [PubMed] [Google Scholar]
- 29.Jeanneret F, Tonoli D, Hochstrasser D, Saurat JH, Sorg O, Boccard J, Rudaz S. Evaluation and identification of dioxin exposure biomarkers in human urine by high-resolution metabolomics, multivariate analysis and in vitro synthesis. Toxicol Lett. 2016;240(1):22–31. doi: 10.1016/j.toxlet.2015.10.004. [DOI] [PubMed] [Google Scholar]
- •30.Perdew GH, Babbs CF. Production of Ah receptor ligands in rat fecal suspensions containing tryptophan or indole-3-carbinol. Nutr Cancer. 1991;16(3–4):209–18. doi: 10.1080/01635589109514159. Primary evidence that the gut microbiota are capable of producing AHR ligands from dietary sources. [DOI] [PubMed] [Google Scholar]
- 31.Zelante T, Iannitti RG, Fallarino F, Gargaro M, De Luca A, Moretti S, Bartoli A, Romani L. Tryptophan Feeding of the IDO1-AhR Axis in Host-Microbial Symbiosis. Front Immunol. 2014;5:640. doi: 10.3389/fimmu.2014.00640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lamas B, Richard ML, Leducq V, Pham HP, Michel ML, Da Costa G, Bridonneau C, Jegou S, Hoffmann TW, Natividad JM, Brot L, Taleb S, Couturier-Maillard A, Nion-Larmurier I, Merabtene F, Seksik P, Bourrier A, Cosnes J, Ryffel B, Beaugerie L, Launay JM, Langella P, Xavier RJ, Sokol H. CARD9 impacts colitis by altering gut microbiota metabolism of tryptophan into aryl hydrocarbon receptor ligands. Nat Med. 2016;22(6):598–605. doi: 10.1038/nm.4102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- •33.Innocentin YLS, Withers DR, Roberts NA, Gallagher AR, Grigorieva EF, Wilhelm C, Veldhoen M. Exogenous Stimuli Maintain Intraepithelial Lymphocytes via Aryl Hydrocarbon Receptor Activation. Cell. 2011;147(3):629–640. doi: 10.1016/j.cell.2011.09.025. First evidence that AHR activation and genotype can influence the microbiota community structure. [DOI] [PubMed] [Google Scholar]
- 34.Maurice CF, Turnbaugh PJ. Quantifying and identifying the active and damaged subsets of indigenous microbial communities. Methods Enzymol. 2013;531:91–107. doi: 10.1016/B978-0-12-407863-5.00005-8. [DOI] [PubMed] [Google Scholar]
- 35.Maurice CF, Turnbaugh PJ. Quantifying the metabolic activities of human-associated microbial communities across multiple ecological scales. FEMS Microbiol Rev. 2013;37(5):830–48. doi: 10.1111/1574-6976.12022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Farmahin R, Crump D, O’Brien JM, Jones SP, Kennedy SW. Time-dependent transcriptomic and biochemical responses of 6-formylindolo [3,2-b]carbazole (FICZ) and 2,3,7,8-tetrachlorodibenzo-p-dioxin (TCDD) are explained by AHR activation time. Biochem Pharmacol. 2016;115:134–43. doi: 10.1016/j.bcp.2016.06.005. [DOI] [PubMed] [Google Scholar]
- 37.Wheeler JL, Martin KC, Resseguie E, Lawrence BP. Differential consequences of two distinct AhR ligands on innate and adaptive immune responses to influenza A virus. Toxicol Sci. 2014;137(2):324–34. doi: 10.1093/toxsci/kft255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nault R, Forgacs AL, Dere E, Zacharewski TR. Comparisons of differential gene expression elicited by TCDD, PCB126, betaNF, or ICZ in mouse hepatoma Hepa1c1c7 cells and C57BL/6 mouse liver. Toxicol Lett. 2013;223(1):52–9. doi: 10.1016/j.toxlet.2013.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- •39.Sato S, Shirakawa H, Tomita S, Ohsaki Y, Haketa K, Tooi O, Santo N, Tohkin M, Furukawa Y, Gonzalez FJ, Komai M. Low-dose dioxins alter gene expression related to cholesterol biosynthesis, lipogenesis, and glucose metabolism through the aryl hydrocarbon receptor-mediated pathway in mouse liver. Toxicol Appl Pharmacol. 2008;229(1):10–9. doi: 10.1016/j.taap.2007.12.029. Primary example that environmental contaminants significantly and permanently alter the microbiome in frog larvae. Provides compelling evidence that similar effects might be observed in other species. [DOI] [PubMed] [Google Scholar]
- 40.Bokulich NA, Chung J, Battaglia T, Henderson N, Jay M, Li H, DLA, Wu F, Perez-Perez GI, Chen Y, Schweizer W, Zheng X, Contreras M, Dominguez-Bello MG, Blaser MJ. Antibiotics, birth mode, and diet shape microbiome maturation during early life. Sci Transl Med. 2016;8(343):343ra82. doi: 10.1126/scitranslmed.aad7121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kohl KD, Cary TL, Karasov WH, Dearing MD. Larval exposure to polychlorinated biphenyl 126 (PCB-126) causes persistent alteration of the amphibian gut microbiota. Environ Toxicol Chem. 2015;34(5):1113–8. doi: 10.1002/etc.2905. [DOI] [PubMed] [Google Scholar]