Abstract
Induced pluripotent stem cells (iPSCs), which are used to produce transplantable tissues, may particularly benefit older patients, who are more likely to suffer from degenerative diseases. However, iPSCs generated from aged donors (A-iPSCs) exhibit higher genomic instability, defects in apoptosis and a blunted DNA damage response compared with iPSCs generated from younger donors. We demonstrated that A-iPSCs exhibit excessive glutathione-mediated reactive oxygen species (ROS) scavenging activity, which blocks the DNA damage response and apoptosis and permits survival of cells with genomic instability. We found that the pluripotency factor ZSCAN10 is poorly expressed in A-iPSCs and addition of ZSCAN10 to the four Yamanaka factors (OCT4, SOX2, KLF4 and c-MYC) during A-iPSC reprogramming normalizes ROS–glutathione homeostasis and the DNA damage response, and recovers genomic stability. Correcting the genomic instability of A-iPSCs will ultimately enhance our ability to produce histocompatible functional tissues from older patients’ own cells that are safe for transplantation.
Induced pluripotent stem cells (iPSCs) hold enormous potential for generating histocompatible transplantable tissue using a patient’s own somatic cells. While older patients are more likely to suffer from degenerative diseases and would benefit from iPSC-based therapies, both basic1–3 and clinical2,4–7 researchers have reported mitochondrial and genomic mutations or instability of iPSCs generated from aged donor tissue (A-iPSCs). In a recent clinical trial of A-iPSCs for age-related macular degeneration (AMD), A-iPSCs generated from one patient donor were found to have genomic instability and were not differentiated to retinal pigment epithelium for transplantation due to concerns about the function and safety of the tissues generated from these cells4,5,7. Therefore, identifying the mechanisms that lead to genomic instability in A-iPSCs and correcting them is imperative for the clinical use of iPSC-based therapies in older patients.
Recent genomics and proteomics analyses have revealed a significant biological role of reactive oxygen species (ROSs) in many intra- and intercellular processes8, from gene expression and protein synthesis to signalling pathways that direct cellular metabolism, chromatin remodelling, the cell cycle, DNA repair and tissue differentiation9. ROS activity has been linked to the cellular aging process10, stem cell fate9, cancer progression11 and multiple diseases, including insulin resistance, diabetes mellitus, cardiovascular disease and neurodegenerative disease12. However, several studies have also identified a protective role of ROSs in cellular processes that are necessary for survival, such as eliminating damaged cells and activation of immune defence responses12. This suggests that organisms must maintain a tight balance of this highly reactive molecule.
Glutathione is a scavenger metabolite for ROSs, and homeostasis of glutathione and ROSs is important to maintain genomic stability13,14. Loss of the homeostatic balance with lower glutathione causes an excess of ROSs, which directly damages DNA. Conversely, excessive glutathione depletes ROSs, which can lead to genomic instability because ROSs are an important cellular signal of stress that induces the DNA damage response. Aberrant ROS depletion therefore increases cell exposure to additional genotoxic stresses, and leads to accumulation of mutations15,16.
Here, we investigated the role of ROS homeostasis in maintaining genomic stability in pluripotent stem cells. We describe the discovery of one mechanism that contributes to A-iPSC instability and a tool—ZSCAN10—that helps protect genomic stability by controlling the homeostatic balance between ROSs and glutathione. We examined this mechanism in iPSCs derived from young and aged mouse donors with the same genetic background and fixed laboratory living conditions, and then extended our work to humans. Understanding how regulation of the ROS and glutathione pathway controls genomic stability in A-iPSCs is highly relevant not only to the therapeutic application of stem cells for age-related diseases but also to the study of the biological role of ROSs in various human diseases.
RESULTS
A-iPSCs show impaired genomic integrity and defects in apoptosis and the DNA damage response compared with Y-iPSCs and ESCs, which are recovered by ZSCAN10 expression
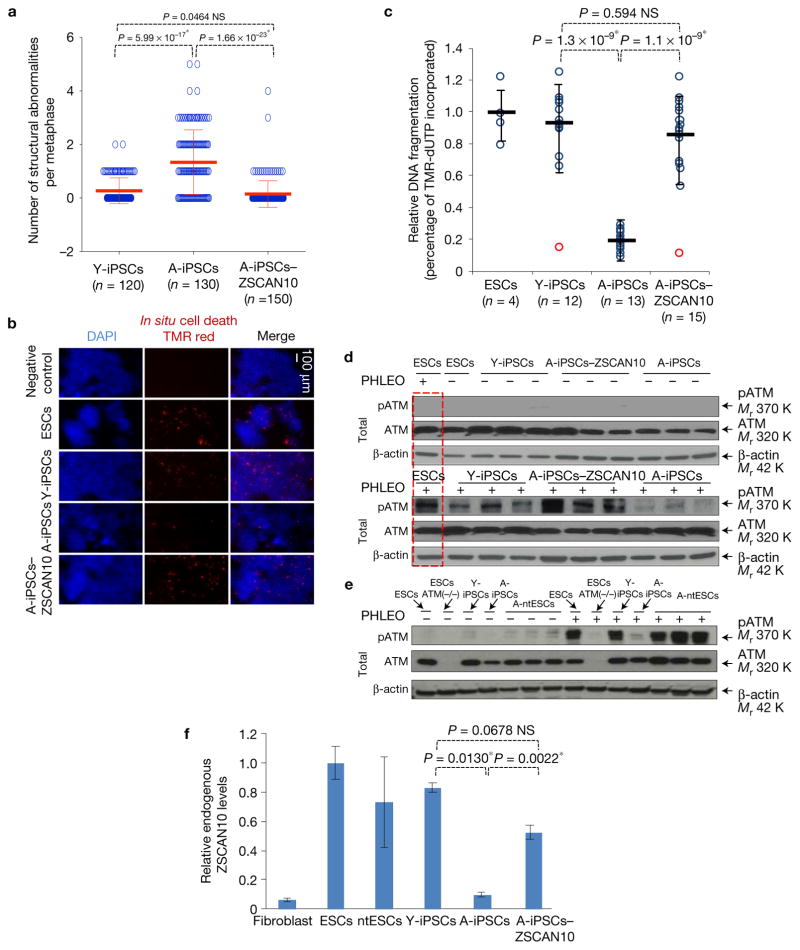
We generated iPSCs from younger donors (Y-iPSCs) (using mouse skin fibroblasts from E17.5 embryos to 5-day-old neonates) and A-iPSCs (using mouse skin fibroblasts from 1.5-year-old adults) as described previously17. We randomly selected a minimum of 12 iPSC clones to undergo a series of common pluripotency tests previously used to characterize mouse and human iPSCs (Supplementary Fig. 1a–e and Supplementary Table 1a)18,19. We also carried out a quantitative PCR (qPCR) analysis of these clones to confirm silencing of the reprogramming factors (Supplementary Fig. 1f). All clones passed the panel of pluripotency tests; however, cytogenetic analysis revealed a greater number of chromosomal structural abnormalities in A-iPSCs (n = 130) compared with Y-iPSCs (n = 120) (Fig. 1a and Supplementary Fig. 1g,h).
Figure 1.
Impaired genomic integrity and DNA damage response of mouse A-iPSCs compared with Y-iPSCs and ESCs, and recovery following transient expression of ZSCAN10. (a) Structural abnormalities observed by cytogenetic analysis in each A-iPSC clone, and recovery with ZSCAN10 expression. The error bars indicate s.e.m. of independent clones analysed per group. Numbers n represent individual metaphases, values are provided in the figure and source data are in Supplementary Table 5. Statistical significance by two-sided t-test followed by post hoc Holm–Bonferroni correction for a significance level of 0.05, * indicates significant and NS not significant. (b) In situ cell death assays of ESCs, Y-iPSCs, A-iPSCs and A-iPSCs–ZSCAN10 were carried out 15 h after phleomycin treatment (PHLEO; 2 h, 30 μg ml−1). A-iPSCs show less staining for cell death. The negative control is Y-iPSCs treated with dye in the absence of enzymatic reaction. The scale bar indicates 100 μm. DAPI, 4,6-diamidino-2-phenylindole. (c) Quantification by image analysis of apoptotic response by DNA fragmentation assay. The error bars indicate s.e.m. of technical and biological replicates. The number of biological replicates is indicated below each group in the figure. Statistical significance by two-sided t-test followed by post hoc Holm–Bonferroni correction for a significance level of 0.05; * indicates significant and NS not significant (Supplementary Table 5). (d) Reduced pATM in A-iPSCs as monitored by immunoblot after phleomycin treatment (2 h, 30 μg ml−1), and recovery of ATM activation following ZSCAN10 expression. The red box indicates the same ESC sample loaded in both immunoblots as an internal control. (e) pATM immunoblot illustrating the differential DNA damage response of A-ntESCs and A-iPSCs generated from an aged tissue donor. Three independent clones of A-ntESCs show a normal DNA damage response after phleomycin treatment. (f) qPCR of ZSCAN10 mRNA levels showing poor activation of ZSCAN10 expression in A-iPSCs and complete activation with transient expression of ZSCAN10. Endogenous ZSCAN10 levels normalized to β-actin. Error bars indicate s.e.m. of two replicates with three independent clones in each sample group (n = 6). Statistical significance by two-sided t-test followed by post hoc Holm–Bonferroni correction for a significance level of 0.05; * indicates significant and NS not significant. Unprocessed original scans of blots are shown in Supplementary Fig. 7.
We also noticed that A-iPSCs showed better survival following manipulative stress, such as passaging and thawing, compared with Y-iPSCs or embryonic stem cells (ESCs). Therefore, we compared the tolerances of mouse A-iPSCs, Y-iPSCs and ESCs to environmental stress (DNA damage). In an in situ cell death assay (Fig. 1b,c), Y-iPSCs (n = 12) and ESC controls (n = 4) showed a significant level of apoptosis after treatment with phleomycin (a structural analogue of bleomycin with higher potency). In contrast, A-iPSCs (n = 13) showed a poorer apoptotic response to phleomycin compared with either Y-iPSCs or ESCs. This observation suggested that a defect in the apoptotic response to DNA damage in A-iPSCs would result in a greater number of cells with genetic abnormalities, reflecting a defect in the elimination of damaged cells20–22.
To define the mechanism underlying the defective apoptotic response in A-iPSCs, we carried out a comparative molecular analysis of three independent clones of A-iPSCs, Y-iPSCs and ESCs, unless specified otherwise in the figures. Compared with Y-iPSCs or ESCs, A-iPSCs consistently exhibited poor activation of the ATM pathway (Fig. 1d), suggesting that the normal cellular mechanisms involved in the DNA damage response are attenuated in A-iPSCs, leading to a failure to eliminate cells with aberrant genomic content. We further demonstrated that A-iPSCs generated from two additional tissue types (lung and bone marrow) exhibited similar defects in the DNA damage response and gene expression patterns (Supplementary Fig. 1i,j).
Nuclear transfer is an alternative reprogramming method to create patient-specific pluripotent stem cells (ntESCs)23. We generated mouse ntESCs by inserting nuclei from aged tissue donors into enucleated oocytes to produce A-ntESCs19,24. Unlike A-iPSCs, the A-ntESCs showed a normal DNA damage response (Fig. 1e) with a normal cytogenetic signature (three clones; Supplementary Fig. 1g). Because oocytes probably contain other reprogramming factors in addition to the four Yamanaka factors used to generate iPSCs, we hypothesized that additional pluripotency factors—present in the enucleated oocyte but absent from aged somatic cells—are required for a normal DNA damage response. We expected that such factors would also be present in Y-iPSCs and ESCs because they have a normal DNA damage response.
Few approaches are available to identify reprogramming factors in enucleated oocytes; however, ESCs were shown to have a similar reprogramming capacity when fused with somatic cells25. This is the same rationale that Yamanaka’s group used to find the four iPSC reprogramming factors in young donor tissue17. We revisited this approach to find additional pluripotency factors present in Y-iPSCs or ESCs but not in A-iPSCs (Supplementary Fig. 2a). A previous study identified 59 core regulators of the pluripotency regulatory network26. We hypothesized that A-iPSCs lack some of these core regulators, which are responsible for the observed poor DNA damage response and genomic instability. To narrow down the candidates, we compared the core genes with genes that are differentially expressed in A-iPSCs versus Y-iPSCs or ESCs. This narrowed down the list to eight candidate genes. We cross-referenced each candidate gene associated with the DNA damage response and genomic stability, and identified the top candidate, ZSCAN10, which had previously been found to functionally associate with ATM, GSS, p53, PARP, PLK1 and ZSCAN4 (refs 27–30). ZSCAN10, a known zinc finger transcription factor, is specifically expressed in ESCs, and has a conserved function in both mouse and human (Supplementary Fig. 2a,b). ZSCAN10 is an integrated part of the transcriptional regulatory network with SOX2, OCT4 and NANOG27,28. We inserted tdTomato reporter into the endogenous ZSCAN10 locus of fibroblasts and carried out time-lapse imaging of fibroblast reprogramming to monitor ZSCAN10 expression31. ZSCAN10 expression was detectable starting on day 6 of reprogramming and was strongly expressed at the time iPSC colonies were formed (Supplementary Fig. 3a).
Endogenous ZSCAN10 expression was high in Y-iPSCs and ESCs, but low in A-iPSCs (Fig. 1f and Supplementary Fig. 3b). Expression of ZSCAN10 with a doxycycline-inducible promoter in A-iPSCs during reprogramming days 5 to 14 (A-iPSCs–ZSCAN10) persistently increased endogenous ZSCAN10 expression to levels similar to those in Y-iPSCs and ESCs (Fig. 1f) A-iPSCs–ZSCAN10 (n = 150) had a reduced number of chromosomal structure abnormalities (Fig. 1a), comparable to the frequency seen in Y-iPSCs and ESCs. A-iPSC–ZSCAN10 clones also showed recovery of apoptosis (Fig. 1b,c) and the DNA damage response (Fig. 1d).
Extended characterization of the DNA damage response in A-iPSCs and recovery by ZSCAN10 expression
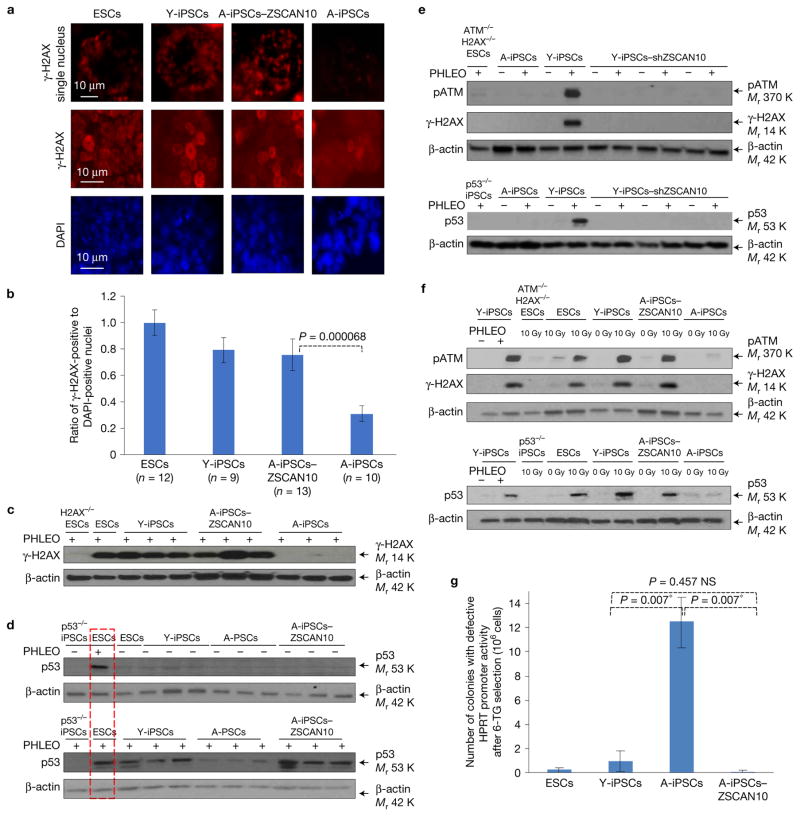
Compared with ntESCs, Y-iPSCs or ESCs, A-iPSCs consistently exhibited poor activation of factors downstream of the ATM pathway (H2AX and p53) (Fig. 2a–d and Supplementary Fig. 3c,d). This recovery was not due to a slower DNA damage response, slower growth rate or differential telomere length in A-iPSCs–ZSCAN10 compared with A-iPSCs (Supplementary Fig. 3e–h). Conversely, downregulation of ZSCAN10 through short hairpin RNA (shRNA) during reprogramming in Y-iPSCs impaired the DNA damage response (Fig. 2e and Supplementary Fig. 3i).
Figure 2.
Evaluation of ZSCAN10 function in DNA damage response and genomic stability of mouse A-iPSCs compared with Y-iPSCs and ESCs. (a) Immunohistochemistry showing low γ-H2AX (phosphorylated H2AX) in A-iPSCs after phleomycin treatment (2 h, 30 μg ml−1) and recovery of γ-H2AX signal with ZSCAN10 expression. (b) Ratio of γ-H2AX-positive cells to DAPI-stained nuclei quantified by immunostaining. Only cells showing punctate γ-H2AX foci were counted. Numbers n represent independent colonies and values are provided in the figure. The error bars indicate s.e.m. of independent colonies. Statistical significance was determined by unpaired two-sided t-test (Supplementary Table 5). (c) Immunoblot analysis of γ-H2AX confirms the immunohistochemistry findings. (d) Immunoblot showing impaired p53 DNA damage response in A-iPSCs and recovery with transient expression of ZSCAN10 in three independent clones after phleomycin treatment (2 h, 30 μg ml−1). The red box indicates the same ESC sample loaded in both immunoblots as an internal control. (e) Immunoblot showing impaired ATM/H2AX/p53 DNA damage response in Y-iPSCs with ZSCAN10 shRNA expression in three independent clones after phleomycin treatment (2 h, 30 μg ml−1). (f) ATM/H2AX/p53-mediated DNA damage response after irradiation. ESCs and Y-iPSCs, but not A-iPSCs, show an increase in pATM/γ-H2AX/p53 level after irradiation (10 Gy). The ATM/H2AX/p53 response to irradiation in A-iPSCs is recovered by transient expression of ZSCAN10. (g) Estimation of higher mutation rate in A-iPSCs, and recovery with ZSCAN10 expression. The mutation frequency was estimated by the inactivation of HPRT promoter activity in the presence of 6-thioguanine (6-TG)-mediated negative selection, and confirmed by qPCR. The error bars indicate s.e.m. of three independent clones in each sample group (n= 3). Statistical significance was determined by two-sided t-test followed by post hoc Holm–Bonferroni correction for a significance level of 0.05; * indicates significant and NS not significant. Unprocessed original scans of blots are shown in Supplementary Fig. 7.
Although the majority of Y-iPSC and A-iPSC-ZSCAN10 clones showed a higher apoptotic response compared with A-iPSCs, two outlier clones did not show a restoration of the apoptotic response (Fig. 1c, red circles). We also found that these outlier clones had low ZSCAN10 expression and a defective DNA damage response (Supplementary Fig. 3j,k), providing further evidence that ZSCAN10 is a positive regulator of genomic stability through the induction of apoptosis in response to DNA damage. The defective DNA damage response of A-iPSCs and its restoration by ZSCAN10 were also confirmed in iPSCs exposed to other DNA damaging agents such as radiation and H2O2 (Fig. 2f and Supplementary Fig. 3l). As a control, Y-iPSCs generated from neonatal skin fibroblasts showed a normal DNA damage response (Supplementary Fig. 3m).
Based on these data, we hypothesized that the failure to eliminate A-iPSCs with DNA damage via apoptosis leads to the accumulation of genomic mutations in A-iPSCs compared with either Y-iPSCs or ESCs. We tested the mutagenic potential of ESCs, Y-iPSCs, A-iPSCs and A-iPSCs–ZSCAN10 using the hypoxanthine phosphoribosyl transferase (HPRT) mutation assay (which measures the mutagenic destruction of HPRT promoter activity) in a population of 106 cells32. We confirmed that A-iPSCs had the highest mutagenicity, which was recovered by ZSCAN10 expression (Fig. 2g).
ZSCAN10 restores ROS–glutathione homeostasis in mouse A-iPSCs via reduction of excessively activated GSS
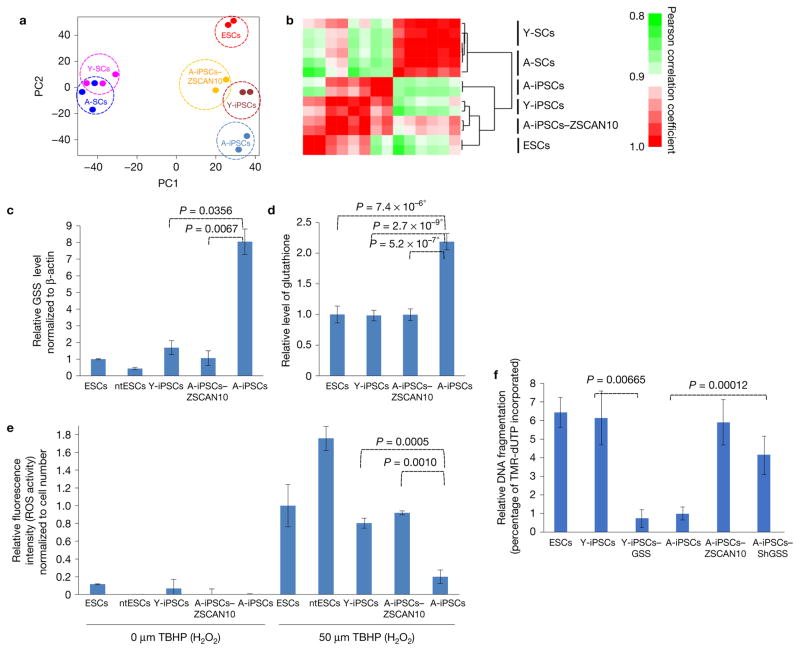
Using supervised clustering analysis, we found that transient expression of ZSCAN10 as a pluripotent transcription factor during reprogramming in A-iPSCs modified the overall pluripotent transcriptional regulatory network to resemble to that of Y-iPSCs (Fig. 3a,b)26. We therefore hypothesized that poor activation of ZSCAN10 during reprogramming of aged donor somatic cells leads to abnormal expression of downstream targets of ZSCAN10, some of which may be involved in the DNA damage response. Indeed, hierarchical clustering analysis of differentially expressed genes within the DNA damage response pathway between A-iPSCs and A-iPSCs–ZSCAN10 demonstrated that expression of ZSCAN10 in A-iPSCs is sufficient to make A-iPSCs–ZSCAN10 cluster with ESCs and Y-iPSCs (Supplementary Fig. 2c). ZSCAN10 target promoters were previously defined in ESCs using chromatin immunoprecipitation (ChIP)-on-chip analysis27. To identify the ZSCAN10 targets involved in the DNA damage response defect in A-iPSCs (Supplementary Fig. 2d), we cross-referenced differentially expressed genes in Y-iPSCs/ESCs and A-iPSCs, genes with altered expression in A-iPSCs compared with A-iPSCs–ZSCAN10, the list of genes with ZSCAN10-targeted promoter binding regions from those previously reported in ChIP-on-chip analysis in ESCs27 and the list of genes extracted from Gene Ontology (GO) involved in the DNA damage response–apoptosis–antioxidant–ROS–genomic stability–cell death pathway. This narrowed down the list to eight candidate genes targeted by ZSCAN10. After we had cross-referenced each candidate gene associated with the DNA damage response and genomic stability, we identified the glutathione regulatory pathway gene, glutathione synthetase (GSS). Because cellular glutathione level is a well-established regulator of DNA damage response and genomic stability13–16, we hypothesized that a glutathione imbalance contributes to the observed defects in A-iPSCs. We carried out hierarchical clustering of differentially expressed genes within the glutathione regulatory pathway (GO:0006749) between A-iPSCs and A-iPSCs–ZSCAN10. We found that A-iPSCs express glutathione regulation-related genes in a distinct pattern, and expression of ZSCAN10 in A-iPSCs is sufficient to make A-iPSCs–ZSCAN10 cluster with ESCs and Y-iPSCs (Supplementary Fig. 2e). GSS was expressed at excessively high levels in A-iPSCs but was downregulated upon ZSCAN10 expression in A-iPSCs, to the levels seen in Y-iPSCs or ESCs (Fig. 3c). Conversely, downregulation of ZSCAN10 by shRNA in Y-iPSCs led to elevated GSS expression (Supplementary Fig. 4a), supporting a role for ZSCAN10 as a suppressor of GSS expression. A-iPSCs generated from two additional tissue types (lung and bone marrow) also exhibited similar expression patterns of ZSCAN10 and GSS (Supplementary Fig. 1i,j). ChIP–qPCR confirmed ZSCAN10 binding activity to the GSS promoter (Supplementary Fig. 4b) to suppress GSS expression (Fig. 3c), as previously reported27.
Figure 3.
Imbalance of ROS–glutathione homeostasis in mouse A-iPSCs, and recovery by ZSCAN10 expression via reduction of GSS. (a,b) Whole-genome expression profiles of aged and young fibroblast cells (A-SCs and Y-SCs) and pluripotent cell lines (ESCs, Y-iPSCs, A-iPSC and A-iPSCs–ZSCAN10) with independent clones for each line as biological repeats (n ≥ 2). (a) Principal component (PC) analysis using whole-genome expression profiles. (b) Heatmap showing hierarchical clustering of samples and pairwise gene expression similarities measured using Pearson correlation coefficient. (c) qPCR of GSS mRNA levels, indicating excessive expression in A-iPSCs and downregulation with ZSCAN10 expression. The error bars indicate s.e.m. of two replicates with three independent clones in each group (n = 6). Statistical significance by two-sided t-test followed by post hoc Holm–Bonferroni correction for a significance level of 0.05 (Supplementary Table 5). (d) High levels of glutathione in A-iPSCs, and recovery by ZSCAN10 expression. Mean ± s.d. is plotted for four biological replicates with two independent clones in each group (n = 8). Statistical significance was determined by two-sided t-test; * indicates significant. (e) ROS scavenging activity. A-iPSCs show strong H2O2 scavenging activity, with reduced response against treatment with tert-butyl hydrogen peroxide (TBHP); the response is recovered by ZSCAN10 expression. Mean ± s.d. is plotted for three independent clones in each group (n = 3). Statistical significance by two-sided t-test (Supplementary Table 5). (f) Apoptosis assay in A-iPSCs with GSS shRNA expression and Y-iPSCs–GSS by image quantification. A lower apoptotic response is seen 15 h after the end of phleomycin treatment (2 h, 30 μg ml−1) in A-iPSCs and in Y-iPSCs–GSS, and is recovered with GSS downregulation in A-iPSCs. The error bars indicate s.e.m. of three biological replicates with two independent clones in each group (n = 6). Statistical significance by two-sided t-test (Supplementary Table 5).
GSS is a key enzyme that drives glutathione synthesis. Glutathione is a scavenger metabolite for ROSs, and homeostatic balance between glutathione and ROSs is important to maintain genomic stability13,14. Loss of the homeostatic balance with lower glutathione causes an excess of ROSs, which directly damages DNA. Conversely, loss of the homeostatic balance with excessive glutathione depletes ROSs, which also induces genomic instability, because ROSs are an important cellular signal of stress that induces the DNA damage response. Improper ROS depletion therefore increases cell exposure to additional genotoxic stresses, and leads to accumulation of mutations15,16. We hypothesized that the low level of ZSCAN10 in A-iPSCs is insufficient to suppress GSS, which leads to excessive glutathione and a loss of glutathione–ROS homeostasis; this in turn depletes the ROS-mediated cellular signal of stress that induces the DNA damage response, and consequently leads to genomic instability. Thus, excessive glutathione in A-iPSCs13 may contribute to the DNA damage response defect and genomic instability.
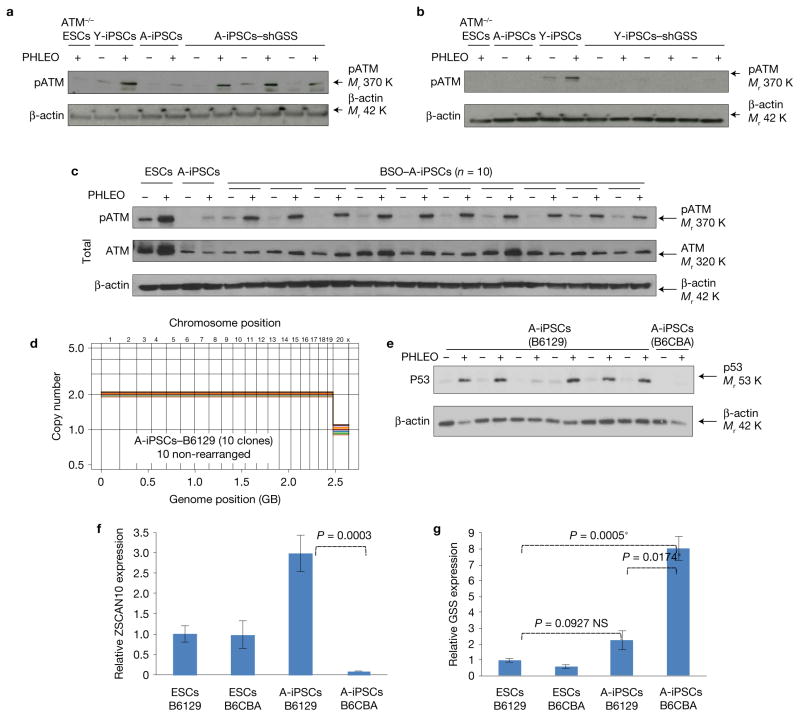
We found that A-iPSCs have excessive levels of glutathione (Fig. 3d) and elevated ROS scavenging activity (Fig. 3e) relative to Y-iPSCs or ESCs. While ROS levels in A-iPSCs were increased by treatment with DNA damaging agents (Fig. 3e) and this might be sufficient to cause direct DNA damage and genomic instability, improper scavenging of ROSs by excess glutathione would limit the ROS cellular stress signal needed to induce the DNA damage response, which would in turn reduce apoptosis and increase A-iPSC exposure to additional genotoxic stress, allowing accumulation of mutations and other genomic alterations. Upon ZSCAN10 expression, glutathione and ROS scavenging activity were normalized to levels equivalent to those seen in Y-iPSCs and ESCs (Fig. 3d,e). In addition, shRNA knockdown of GSS in reprogrammed A-iPSCs (Supplementary Fig. 4c) decreased glutathione levels and ROS scavenging activity (Supplementary Fig. 4d,e), and increased apoptosis (Fig. 3f). Conversely, overexpression of GSS in Y-iPSCs (Supplementary Fig. 4f) increased glutathione and ROS scavenging activity (Supplementary Fig. 4d,e), and decreased apoptosis (Fig. 3f). The impact of changes in GSS expression on DNA damage responses was confirmed with both genetic approaches (Fig. 4a,b) and pharmacological treatment of A-iPSCs with the GSS inhibitor L-buthionine-sulfoximine (BSO) (Fig. 4c). Consistent with our findings, a similar glutathione-mediated inhibition of the DNA damage response has previously been reported in chemotherapy-resistant cancers and in primary lung, pancreatic and colorectal cancers14,33–35. Interestingly, we observed variability in A-iPSCs derived from mice of different genetic backgrounds: more A-iPSC clones from B6129 mice showed genomic stability, with a normal DNA damage response, higher ZSCAN10 expression and lower GSS expression (Fig. 4d–g), compared with A-iPSCs from B6CBA mice (Figs 1 and 3c).
Figure 4.
Evaluation of GSS regulation on DNA damage response and effects of different genetic backgrounds on GSS regulation. (a) Immunoblot of pATM showing recovery of the DNA damage response after phleomycin (PHLEO) treatment in three independent clones of A-iPSCs with shRNA-mediated knockdown of GSS (also see Supplementary Fig. 4c). (b) Immunoblot of pATM showing that expression of GSS complementary DNA (also see Supplementary Fig. 4f) impairs the DNA damage response in three independent clones of Y-iPSCs after phleomycin treatment. (c) Immunoblot of pATM showing recovery of the DNA damage response after phleomycin treatment in 10 independent clones of A-iPSCs with BSO (0.5 mM)-mediated inhibition of GSS. (d) Copy number profiling analysis of A-iPSCs (n = 10) generated from fibroblasts from tail tip skin of a 1.5-year-old B6129 mouse. (e) Immunoblot of p53 showing DNA damage response after phleomycin treatment in the majority of independent clones of A-iPSCs generated from a B6129 mouse. A poor DNA damage response (indicated by a lack of p53 upregulation after phleomycin treatment) was seen less frequently in A-iPSCs from B6129 mouse donors than in A-iPSCs from B6CBA mouse donors. However, the poor DNA damage response clone still shows the minimum induction of the DNA damage response. (f,g) Expression levels of ZSCAN10 and GSS in A-iPSCs from a B6129 mouse. The error bars indicate s.e.m. of two technical replicates with three independent clones in each sample group (n = 3). Statistical significance was determined by two-sided t-test followed by post hoc Holm–Bonferroni correction for a significance level of 0.05; * indicates significant and NS not significant. Unprocessed original scans of blots are shown in Supplementary Fig. 7.
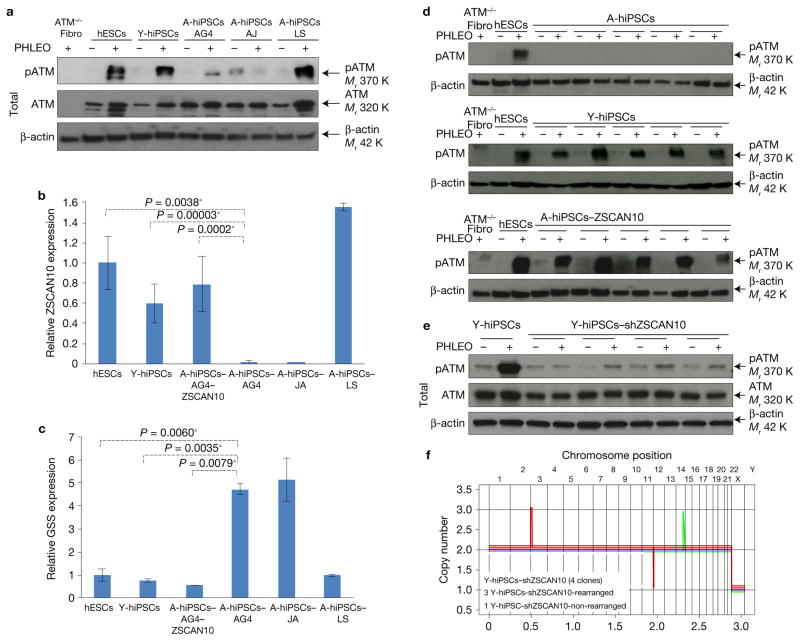
ZSCAN10 recovers the DNA damage response in human A-hiPSCs caused by excessive GSS
To determine whether the mechanisms underlying genomic instability observed in mouse A-iPSCs are also present in humans (cross-species conservation), we first analysed two previously published human A-iPSC clones, A-hiPSC-JA1 and A-hiPSC-LS36, and an unpublished A-hiPSC-AG4 clone from a 71-year-old donor (from Cooperative Human Tissue Network, a National Cancer Institute). Consistent with the phenotypes observed in mouse A-iPSCs, A-hiPSC-JA and A-hiPSC-AG4 showed a poor DNA damage response (Fig. 5a), low levels of ZSCAN10 (Fig. 5b), high levels of GSS (Fig. 5c) and genomic instability (previously reported1). However, A-hiPSC-LS did not exhibit these aging phenotypes and had a normal DNA damage response, normal ZSCAN10/GSS expression (Fig. 5a–c) and genomic stability (previously reported36). Similar clonal variation among human tissue donors was described in the recent A-hiPSC clinical trial for the treatment of AMD4,5,7. In that trial, treatment proceeded successfully with A-hiPSCs generated from the first patient without significant genomic instability, but the trial was halted upon discovery of genomic instability in A-hiPSCs generated from the second patient4,5,7. Together, these observations underscore the idea that, even as we discover mechanisms that contribute to the aging phenotype in A-iPSCs, differences in genetic polymorphisms (and lifestyle) play critical roles in aging and its biological effects on iPSC reprogramming in both mouse and human models. This is a well-established concept in somatic cell aging37.
Figure 5.
Evaluation of ZSCAN10 function in DNA damage response and genomic integrity in human A-hiPSCs. (a) Immunoblots showing the levels of pATM and β-actin proteins with three imported A-hiPSC clones with known abnormal cytogenetic signature. (b) qPCR of ZSCAN10. The error bars indicate s.e.m. of two replicates with three independent clones (n= 6) in each sample group, except three biological replicates (n = 3) in the sample of A-iPSC-JA and A-iPSC-LS. Statistical significance was determined by two-sided t-test followed by post hoc Holm–Bonferroni correction for a significance level of 0.05; * indicates significant. (c) qPCR of GSS. Statistical significance was determined by two-sided t-test followed by post hoc Holm–Bonferroni correction for a significance level of 0.05; * indicates significant. The error bars indicate s.e.m. of two replicates with three independent clones (n = 6) in each sample group, except three biological replicates (n= 3) in the sample of A-iPSC-JA and A-iPSC-LS. (d) Immunoblots showing the levels of pATM and β-actin in five independent clones of A-hiPSCs, five independent clones of Y-hiPSCs and five clones of A-hiPSCs expressing ZSCAN10. (e) Immunoblot showing impaired ATM DNA damage response in Y-hiPSCs with ZSCAN10 shRNA expression in three independent clones after phleomycin treatment (2 h, 30 μg ml−1). (f) Copy number profiling analysis of Y-hiPSCs with ZSCAN10 shRNA expression in four independent clones43. Unprocessed original scans of blots are shown in Supplementary Fig. 7.
To explore the cross-species conservation of the mechanism that maintains ROS–glutathione homeostasis, we utilized AG4 fibroblasts with a confirmed poor DNA damage response, and generated A-hiPSCs in the presence and absence of human ZSCAN10 expression using a doxycycline system38. Each A-hiPSC clone was put through a series of pluripotency tests and compared with hESCs and Y-hiPSCs derived from fibroblasts (Supplementary Fig. 5a,b and Supplementary Table 1b). As we observed in mouse A-iPSCs, endogenous ZSCAN10 expression was significantly lower in A-hiPSCs than Y-hiPSCs or hESCs (Fig. 5b). A-hiPSCs also showed a blunted DNA damage response (ATM phosphorylation, pATM; Fig. 5d) and a poorer apoptotic response to phleomycin (Supplementary Fig. 5c) compared with Y-hiPSCs or hESCs. The poor DNA damage response in A-hiPSCs was confirmed with various reprogramming vectors such as lentivirus reprogramming without MYC39,40 and an integration-free episomal vector system (Supplementary Fig. 5d,e)41, suggesting that the observed phenotype of A-hiPSCs is not caused by reprogramming vector systems or viral vector integration. As with the reprogramming of aged mouse donor cells, transient expression of ZSCAN10 during reprogramming days 5 to 15 in A-hiPSCs (A-hiPSCs–ZSCAN10) persistently increased endogenous ZSCAN10 expression to levels similar to those in Y-hiPSCs and hESCs (Fig. 5b). Importantly, increased ZSCAN10 expression recovered the DNA damage response (Fig. 5d) and the apoptosis defect (Supplementary Fig. 5c) in A-hiPSCs. Also consistent with the mouse data, we found that A-hiPSCs express higher levels of GSS (Fig. 5c), which were normalized by increased expression of ZSCAN10 (Fig. 5c). Conversely, shRNA knockdown of ZSCAN10 in Y-hiPSCs impaired the DNA damage response (Fig. 5e) and genomic stability (Fig. 5f). In addition, shRNA knockdown of ZSCAN10 in hiPSCs generated from a previously reported secondary reprogramming system, in which H1 hESC-derived fibroblasts were reprogrammed into hiPSCs (equivalent to Y-hiPSCs) by pre-integrated doxycycline-inducible reprogramming lentivirus42, impaired the DNA damage response (Supplementary Fig. 5f). ChIP–qPCR confirmed that ZSCAN10 directly binds to the ZSCAN10 DNA binding motif on the human GSS promoter (Supplementary Fig. 5g) to suppress GSS expression (Fig. 5c).
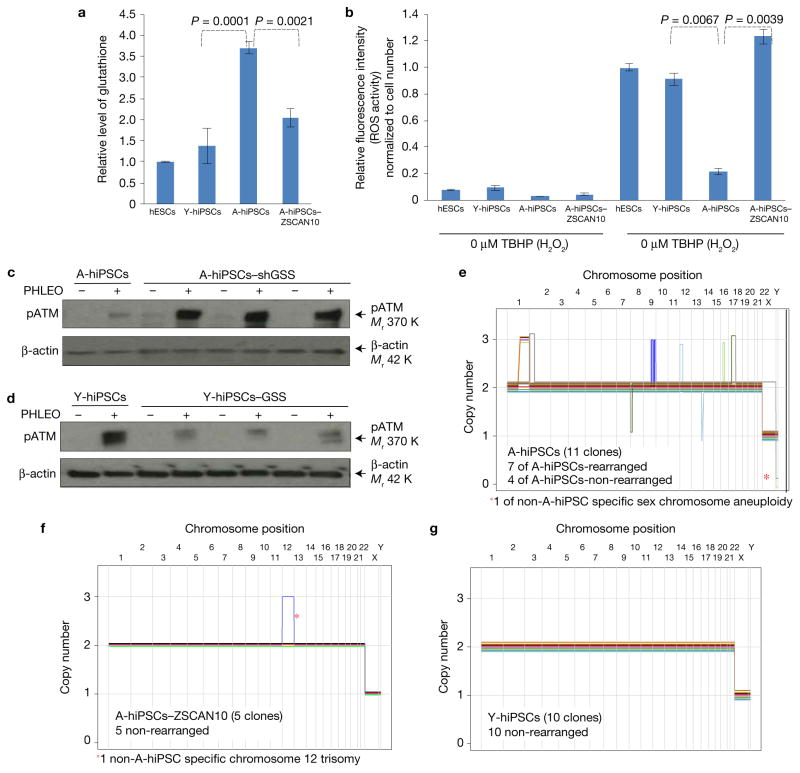
A-hiPSCs had excessive levels of glutathione (Fig. 6a) and elevated ROS scavenging activity (Fig. 6b) relative to Y-hiPSCs or hESCs. On ZSCAN10 expression, glutathione and ROS scavenging activity were normalized to levels equivalent to those seen in Y-hiPSCs and hESCs (Fig. 6a,b). shRNA knockdown of GSS in A-hiPSCs recovered the DNA damage response (Fig. 6c), while overexpression of GSS in Y-hiPSCs blunted the DNA damage response (Fig. 6d). Together, these data confirm the evolutionary conservation of a regulatory mechanism by which ZSCAN10 normalizes GSS levels and ROS–glutathione homeostasis, and recovers the DNA damage response in two mouse and five human cell lines.
Figure 6.
Impaired DNA damage response in human A-hiPSCs caused by deregulation of ZSCAN10 and GSS and recovered by ZSCAN10 expression. (a) Excessive oxidation capacity with elevated glutathione in A-hiPSCs, and recovery by ZSCAN10 expression. The total glutathione level was measured to determine the maximum oxidation capacity. Excessive oxidation capacity of glutathione in A-hiPSCs is normalized to the level of hESCs and Y-hiPSCs by transient expression of ZSCAN10. Glutathione analysis was conducted with the glutathione fluorometric assay. Mean ± s.d. is plotted for three biological replicates with two independent clones (n = 6) in each sample group from each condition. Statistical significance was determined by two-sided t-test. (b) ROS scavenging activity of hESCs, Y-hiPSCs, A-hiPSCs and A-hiPSCs–ZSCAN10. A cellular ROS assay kit (DCFDA assay) was used to measure H2O2 scavenging activity. A-hiPSCs show strong H2O2 scavenging activity, with a reduced response against treatment with TBHP (tert-butyl hydrogen peroxide; stable chemical form of H2O2, 3 h); the response is recovered by ZSCAN10 expression. Mean ± s.d. is plotted for four biological replicates in each sample group from each condition (n= 4). Statistical significance was determined by two-sided t-test. (c) Immunoblot of pATM showing recovery of the DNA damage response after phleomycin treatment in three independent clones of A-hiPSCs with shRNA-mediated knockdown of GSS. (d) Immunoblot of pATM showing that lentiviral expression of GSS cDNA impairs the DNA damage response in three independent clones of Y-hiPSCs after phleomycin treatment. (e–g) Copy number profiling analysis of human iPSCs43. Schematic diagrams represent seven rearranged A-hiPSCs, four non-rearranged A-hiPSCs and five non-rearranged A-hiPSCs–ZSCAN10 in the genetically controlled setting of A-hiPSCs. Ten non-rearranged Y-hiPSCs, which were generated from a different tissue donor, were also included. A-hiPSCs (n = 11 (7/11), P = 0.64), A-hiPSCs–ZSCAN10 (n= 5 (0/5), P* = 6.3× 10−3) and Y-hiPSCs (n= 10 (0/10), P* < 4× 10−5). The number in parentheses represents detected rearrangements and P and P* are the observed and estimated likelihoods of detecting no rearrangements in the absence of lineage effects using a binomial distribution, respectively50. Unprocessed original scans of blots are shown in Supplementary Fig. 7.
ZSCAN10 maintains genomic integrity in human A-hiPSCs
We examined chromosomal structural abnormalities (for example, translocation, duplication and deletion) in A-hiPSC clones by a combination of DNA sequencing-based copy number variation analysis43 and karyotyping analysis to confirm the effect of ZSCAN10 on genomic instability. We found that seven of 11 independent A-hiPSC clones showed a cytogenetic abnormality in eight regions (Fig. 6e), while five A-hiPSCs–ZSCAN10 and 10 Y-iPSCs did not (Fig. 6f and Supplementary Fig. 5h,i). However, we also observed sex chromosome aneuploidy in one A-hiPSC clone and trisomy 12 in one A-iPSC-ZSCAN10 clone, which are common chromosomal alterations in pluripotent stem cell culture and more likely to have been introduced by in vitro expansion and not by A-iPSC-specific reprogramming44,45.
To explore whether mutation rates are altered in A-hiPSCs, we carried out whole exome sequencing analysis in a randomly selected subset of A-hiPSCs (three clones with a normal cytogenetic signature and five clones with cytogenetic alterations), and A-hiPSCs with ZSCAN10 expression (four clones) using the somatic cells as a reference genomic sequence. We found two dominant nonsynonymous point mutations and two synonymous mutations (Supplementary Fig. 5j). The cytogenetic and point mutation analyses revealed that all A-hiPSC clones contain cytogenetic abnormalities or nonsynonymous point mutations, which were not observed in the A-hiPSC–ZSCAN10 clones. Absence of common cytogenetic abnormalities or point mutations in fibroblasts used to generate A-iPSCs was confirmed by karyotyping (screening 20 clones), chromosome painting (screening 100 clones) and whole exome sequencing (80× coverage). Therefore, recurrent cytogenetic abnormalities or point mutations in independent clones of A-hiPSCs may be induced either during iPSC reprogramming or exist at low frequency prior to reprogramming, which would give a selective reprogramming or growth advantage to aged cells. However, ZSCAN10 expression reduced the selective advantage of genomic alterations. In both the mouse and human models (Figs 1a and 2g, and 6e,f), ZSCAN10 expression in A-hiPSCs during reprogramming increased the likelihood of obtaining A-hiPSCs with genomic stability. We also confirmed the cytogenetic stability of Y-hiPSCs–shZSCAN10 (Fig. 5e,f). These human data confirm that the effect of ZSCAN10 on genomic stability is evolutionarily conserved, with ZSCAN10 recovering genomic stability in A-hiPSCs and recapitulating what was seen in the mouse model.
DISCUSSION
We found that the pluripotency factor ZSCAN10 is poorly expressed in A-iPSCs and addition of ZSCAN10 during A-iPSC reprogramming recovers genomic stability by normalizing the homeostatic balance of ROS–glutathione and the DNA damage response. We also confirmed that this mechanism is conserved across species, with the same effects of ZSCAN10 on the DNA damage response and genomic stability in A-iPSCs from mice and humans. Another member of the ZSCAN family, ZSCAN4, may help maintain genomic integrity of Y-iPSCs30; however, ZSCAN4 was abundantly expressed at similar levels in both Y-iPSCs/ESCs and A-iPSCs. Therefore, we excluded ZSCAN4 as a candidate gene involved in the observed age-related defects in A-iPSC DNA damage response and genomic stability, although the present data cannot rule out ZSCAN4–ZSCAN10 synergy in this setting.
The main driver for the poor DNA damage response and genomic instability in A-iPSCs and why some A-iPSCs show a more pronounced aging phenotype are still unclear among the different tissue donors. One possibility is that the main driver for the observed aging phenotypes in A-iPSCs originates in the somatic cells themselves. However, it is challenging to identify a single causal genetic or epigenetic factor for a given aging phenotype among the different tissue donors, considering the complex genetic and epigenetic alterations that occur with aging. Ageing phenotypes are most likely caused by a combination of multiple alterations that accumulate over time. Nevertheless, our data suggest that one driving force for the poor DNA damage response in A-iPSCs could be the elevation of glutathione. iPSC reprogramming may serve as a strong selection of certain cells among the cell population during epigenetic remodelling.
Recent publications2,3,46 have alluded to genomic instability in iPSCs, but the mechanisms causing genomic instability and approaches used to recover it have not been addressed in detail. Here, we have demonstrated that the genomic stability of iPSCs varies for different somatic cell donors. Reference 46 reached similar conclusions, finding that the genetic alterations observed in iPSCs were attributable to differences among the individual donors. We have gone a step further by demonstrating that modulation of the ZSCAN10–GSS pathway during iPSC reprogramming has the potential to restore the DNA damage response and recover genomic stability in both mouse and human cells. Nevertheless, additional research is needed to fully understand whether A-iPSCs generated from different somatic cell types and various somatic cell donors, or using different reprogramming vector systems with additional reprogramming factors, maintain genomic stability in similar ways. Our data demonstrate that A-iPSCs derived from some donors do not exhibit defects in DNA damage responses and genomic stability, which is consistent with the previous finding that genetic differences among donors are a major determinant of clonal variance of iPSCs. It will be of great importance to define what additional genetic or environmental factors contribute to the clonal variance in future studies.
It is likely that a higher ROS level in the somatic cells serves as the selective pressure for higher glutathione levels during iPSC reprogramming. Interestingly, elevation of ROSs is a well-known physiological change that occurs during aging47, and ROS variations in aging cells and individuals has been reported among different tissue donors48. We thus hypothesize that an increase in cellular ROSs that occurs during the normal aging of somatic cells from different tissue donors may drive and sustain the elevation in glutathione seen during A-iPSC reprogramming, leading to genomic instability in the A-iPSCs. In support of this hypothesis, we found that cellular ROS levels (detected by MitoSOX staining) were low in young donor somatic cells (Y-SCs) from B6CBA mice and aged donor somatic cells (A-SCs) from B6129 mice, but high in A-SCs from B6CBA mice (Supplementary Fig. 6A). This variability among the different tissue donors was also observed in human somatic cells, with low cellular ROS levels in Y-SCs from donor MRC5 and A-SCs from donor LS, but high in A-SCs from donor AG4 (Supplementary Figs 6C and 6D). Interestingly, the cellular ROS level among the different tissue donors was highly correlated with a poor DNA damage response/genomic instability in the reprogrammed iPSCs (Supplementary Figs 6E and 6F). To investigate the direct effect of the cellular ROS reduction on the recovery of DNA damage response/genomic stability in the reprogrammed A-iPSCs, we used the stabilized form of glutathione (glutathione reduced ethyl ester), as previously reported49, to reduce cellular ROS levels (Supplementary Figs 6B and 6D). We hypothesized that the reduction of ROSs by treating aged somatic cells with glutathione prior to and during the early stage of reprogramming may remove the selective advantage for elevated glutathione, overcome the reprogramming barrier and improve genomic stability in A-iPSCs. We examined the effect of glutathione reduced ethyl ester treatment of human AG4 fibroblast clones, which show a higher level of MitoSOX staining, prior to and during the first 10 days of A-hiPSC reprogramming. The results suggest that treatment protects the DNA damage response (Supplementary Fig. 6G) and maintains genomic stability (upper panel, Supplementary Fig. 6H) compared with untreated A-hiPSC (lower panel, Supplementary Fig. 6H; P = 0.00005). In addition, ZSCAN10 levels in A-iPSCs were elevated after glutathione reduced ethyl ester treatment (Supplementary Fig. 6I), indicating that glutathione treatment also influences epigenetic changes and pluripotent gene expression during iPSC reprogramming. GSS levels in A-iPSCs were reduced with glutathione reduced ethyl ester treatment (Supplementary Fig. 6J). While the treatment with stabilized glutathione was carried out in one AG4 donor line, this set of experiments suggest that reduction of ROSs early in reprogramming essentially removed the force in the somatic cells (age-related ROS elevation among the different tissue donors) that was driving the elevated GSS expression in A-iPSCs.
Because disruption of ROS–glutathione homeostasis has been associated with human diseases such as cancer, diabetes and cardiovascular disorders, our discovery has broader implications for the pathobiology of these conditions12. Understanding how regulation of the ROS–glutathione pathway modulates the apoptotic signal in cells, thereby impacting genomic stability, is highly relevant not only to research in aging and the therapeutic application of stem cells, but also to research into cancer biology and therapeutics.
To realize the benefits of hiPSCs as a source of histocompatible transplantable tissue, particularly for older individuals who stand to benefit the most from iPSC-based regenerative medicine, a full understanding of the factors that drive the aging phenotype in A-iPSCs is needed. Ultimately, this work will lead to the development of innovative tools for producing safer patient-specific stem cells for older patients.
METHODS
Cell culture
Mouse ESCs and iPSCs were cultured in ESC medium containing 15% fetal bovine serum (FBS) and 1,000 U ml−1 of leukaemia inhibitory factor (ESGRO LIF, 1 million units 1 ml−1). Mouse ESCs were generated and their pluripotency was tested as reported in our previous publication19. Doxycycline-inducible ZSCAN10 was induced in medium supplemented with 2 μg ml−1 doxycycline (MP Biomedicals, doxycycline hyclate). Human iPSCs were cultured in human ESC medium (DMEM/F12, 20% knockout serum replacement, 10 ng ml−1 basic fibroblast growth factor and 0.1 mM β-mercaptoethanol) on irradiated mouse embryonic feeders. Human and mouse fibroblasts were cultured in DMEM with 10% FBS.
Generation of Y-iPSCs, A-iPSCs, A-iPSCs–ZSCAN10 and Y-iPSCs–shZSCAN10
Fibroblasts were collected from B6CBAF1 mouse E17.5 embryonic skin, 5-day-old tail tip skin and 1.5-year-old tail tip skin. Fibroblasts from B6129 mice were collected from 1.5-year-old tail tip skin. Fibroblasts were not cultured for more than three passages to limit extensive in vitro culture. 106 fibroblasts were infected with retrovirus generated from pMX-mOCT4, pMX-mSOX2, pMX-mKLF4 and pEYK-mMYC in six-well dishes with 0.5 ml of each viral supernatant (total 2 ml per well) and spun at 2,500 r.p.m. at room temperature for 90 min (benchtop centrifuge, Beckman Coulter, Allegra 6R). For the generation of A-iPSCs–ZSCAN10, the procedure was identical, but in addition to the four reprogramming factors we added a doxycycline-inducible system to overexpress ZSCAN10. This system consisted of two lentiviruses generated from a plentiRZ–ZSCAN10 and a plenti-RTTA vector. For the generation of Y-iPSCs–shZSCAN10, the procedure was identical, but in addition to the four reprogramming factors we added a set of shRNA viruses for ZSCAN10 (4 GIPZ lentiviral shRNA vectors from abmgood.com: NM_001033425.3). All cells infected with the reprogramming factors and those with additional ZSCAN10 were plated on irradiated CF-1 mouse embryonic feeder cells in a 10 cm tissue culture dish in ESC medium containing 20% FBS and 1,000 U ml−1 of LIF. Media were changed on day 2 and doxycycline addition started on day 3 to induce ZSCAN10 overexpression. Floating cells were collected by medium centrifugation and returned to culture during media changes. On day 4, cultured cells were trypsinized and replated onto four 10 cm dishes pre-coated with gelatin (0.1%) and irradiated mouse embryonic fibroblasts (MEFs) in ESC maintenance medium. Media were changed daily until ESC-like colonies were observed. The reprogrammed colonies were tested for pluripotency based on teratoma formation, alkaline phosphatase staining, SSEA-1 and NANOG staining and OCT4 expression levels.
Generation of ZSCAN10 Tomato fluorescence protein reporter by CRISPR targeting
The 5′ end of the ZSCAN10 starting codon region in ESCs was targeted to insert the Tomato fluorescent protein with the CRISPR genomic targeting tool. Briefly, the left and right arms of mZSCAN10 were cloned into the plasmid PCR2.1 IRES TOMATO P2A flagbio (Invitrogen) using the following primer sets.
LEFT-ZSCAN10-nhe-F: 5′-attgctagcGAGGACTACTTGTGGAAGTCAG TG-3′. LEFT-ZSCAN10-bamh-R: 5′-attggatccggatccttgggagaattcaggg-3′. RIGHT-ZSCAN10-NOT-F: 5′-attGCGGCCGCatgctggcggaaccagtccc-3′. RIGHT-ZSCAN10-ASC-R: 5′-attGGCGCGCCacagacagattggacagccaggac-3′. The resulting plasmid, along with guide RNA 5′-CACCgATACTGCGTTAAGATCTGAC-3′, 5′-aaacGT CAGATCTTAACGCAGTATc-3′, 5′-CACCgTTTAGCTCCACAGGTGCAGG-3′, 5′-aaacCCTGCACCTGTGGAGCTAAAc-3′ and CAS 9, were transfected into ESCs, and individual clones were screened for integration by PCR and DNA sequencing.
Generation of L-A-iPSCs and BM-A-iPSCs
Whole tissue from lung (106) and bone marrow (106) was collected from 1.4-year-old mice. The lung tissues were dissociated with collagenase (1 mg ml−1) first and then mechanically, and cells were plated in 10 cm3 dishes, where they were cultured in DMEM with 20% serum. The cells were directly infected with iPSC reprogramming viruses as described above. The resulting cells were cultured in DMEM for 4 days and then transferred to ESC maintenance medium on MEFs. Bone marrow tissue was treated to lyse the red blood cells and then was plated on mitomycin-treated OP9 cells in IMDM (Invitrogen) supplemented with 10% FBS, 1× penicillin/streptomycin/glutamine (Invitrogen), vascular endothelial growth factor (R&D Systems, 40 ng ml−1), FLT (R&D Systems, 100 ng ml−1), thrombopoietin (R&D Systems, 100 ng ml−1) and stem cell factor (R&D Systems, 40 ng ml−1) on day 0. The cells were directly infected with iPSC reprogramming viruses as described above. The resulting cells were cultured in DMEM for 4 days and then transferred to ESC maintenance medium on MEFs.
Generation of BSO-A-iPSCs
BSO-A-iPSCs were generated in the presence of 500 μM of BSO (Sigma, B2515) starting at the end of reprogramming day 5.
Generation of human iPSCs
For the generation of human A-hiPSCs and Y-hiPSCs, 105 skin fibroblasts (AG4) from a 71-year-old individual and MRC5 fibroblasts (human fetal fibroblast cells) at passages 2–7 were plated in gelatin-coated six-well plates at a density of 1 × 105 cells per well and transduced 24 h later with the retroviral tetracistronic vector (SFG-SV2A) encoding all four Yamanaka reprogramming factors (OCT4, SOX2, KLF4 and c-MYC) in a single transcript in the presence of 4 μg ml−1 of polybrene. Media were changed 48 h later and replaced every day thereafter with human ESC medium (DMEM/F12, 20% knockout serum replacement, 10 ng ml−1 basic fibroblast growth factor and 0.1 mM β-mercaptoethanol). 15–25 days after transduction, colonies with hESC morphology were mechanically dissociated and transferred into plates pre-seeded with irradiated MEFs (GlobalStem). Cells were thereafter passaged with dispase and expanded to establish iPSC lines. For the generation of A-hiPSCs–ZSCAN10, the procedure was identical, but in addition to the four reprogramming factors we added a doxycycline-inducible system to overexpress ZSCAN10 as described above for mouse iPSCs. All cells infected with the reprogramming factors and those with additional ZSCAN10 were plated on irradiated CF-1 mouse embryonic feeder cells in a 10 cm tissue culture dish in human ESC medium (DMEM/F12, 20% knockout serum replacement, 10 ng ml−1 basic fibroblast growth factor and 0.1 mM β-mercaptoethanol). Media were changed on day 2 and doxycycline addition started on day 3 to induce ZSCAN10 overexpression. Floating cells were collected by medium centrifugation and returned to culture during media changes. On day 4, cultured cells were trypsinized and replated onto four 10 cm dishes pre-coated with gelatin (0.1%) and irradiated MEFs in ESC maintenance medium. Media were changed daily until ESC-like colonies were observed. The source of the cell lines and the methods used for identification and authentication are summarized in Supplementary Table 1.
For the generation of A-hiPSCs–glutathione, we pre-treated the AG4 fibroblasts by adding 3 mM glutathione reduced ethyl ester (GOLDBIO, catalogue no G-275-500) to the media and continued the treatment until day 10 of reprogramming.
Fibroblasts used to generate human A-iPSCs (AG4) were from CHTN (Cooperative Human Tissue Network, a National Cancer Institute. http://www.chtn.org). We also imported human A-iPSC clones from the laboratory of C.-W.L (clone AG4; Rutgers University, New Brunswick, NJ, USA; 71 years of age), J. Adjaye’s laboratory (A-hiPSC-JA; Max Planck Institute for Molecular Genetics, Berlin, Germany; 84 years of age)1, and L. Studer (A-hiPSC-LS; Sloan Kettering Institute, New York, USA; 82 years of age)36. MRC5 fibroblasts used to generate human young iPSCs were imported from ATCC. Human pluripotent stem cell research is compliant with all relevant ethical regulations by the tri-institutional stem cell initiative of Weill Cornell Medical College, Rockefeller University and Memorial Sloan Kettering Cancer Center (approval number 2013-09-002).
Retrovirus generation
293T cells were seeded overnight at 5 × 106 cells per 150 mm dish with DMEM supplemented with 10% FBS and penicillin/streptomycin. Retrovirus was generated using pMX-mOCT4, pMX-mSOX2, pMX-mKLF4 and pEYK-mMYC or the tetracistronic SFG-SV2A vector encoding hOCT4, hSOX2, hKLF4 and hMYC constructs as described previously. The cells were transfected with calcium phosphate. Media were replaced with fresh DMEM twice, 18 h after transfection. Approximately 48 h after transfection, medium containing the retrovirus was collected and the cellular debris was removed by centrifugation. The supernatant was filtered through a 0.45 μm filter, and the retrovirus was pelleted with ultracentrifugation at 33,000 r.p.m. in a 45Ti rotor (Beckman) for 90 min at 4 °C. The retroviral particles were resuspended in ESC medium and stored at −80 °C.
Lentivirus production
293T cells were seeded overnight at 5× 106 cells per 150 mm dish with DMEM supplemented with 10% FBS and penicillin/streptomycin. The cells were transfected with plentiRZ–ZSCAN10 (mouse and human), plenti-RTTA or plenti-GSS, with calcium phosphate cell transfection, as previously described51. The ZSCAN10 cDNA was clone MmCD00295052 in the pENTR223.1 backbone and the GSS cDNA was clone MmCD00313013 in the pCMV SPORT6 backbone from the Harvard Plasmids core (http://plasmid.med.harvard.edu/PLASMID/Home.jsp). The cDNA for mouse and human ZSCAN10 was subcloned into a plentiRZ vector. At 48 h after transfection, the medium containing the lentivirus was collected and the cellular debris was removed by centrifugation. The supernatant was filtered through a 0.45 μm filter, and the lentivirus was pelleted by ultracentrifugation at 33,000 r.p.m. in a 45Ti rotor (Beckman) for 90 min at 4 °C. The lentivirus particles were resuspended in DMEM and stored at −80 °C.
Quantitative real-time PCR (qPCR) analysis
The expression levels of pluripotency genes (ZSCAN10, OCT4, GSS and β-actin) were quantified by qPCR. Total RNA (1 μg) was reverse transcribed in a volume of 20 μl using the M-MuLV reverse transcriptase system (New England Biolabs), and the resulting cDNA was diluted into a total volume of 200 μl. 10 μl of this synthesized cDNA solution was used for analysis. For pluripotency genes, each reaction was carried out in a 25 μl volume using the Power SYBR Green PCR Mastermix (Applied Biosystems). The conditions were programmed as follows: initial denaturation at 95 °C for 10 min followed by 40 cycles of 30 s at 95 °C, 1 min at 55 °C and 1 min at 72 °C; then 1 min at 95 °C, 30 s at 55 °C and 30 s at 95 °C. All of the samples were duplicated, and the PCR reaction was carried out using a Mx3005P reader (Stratagene), which can detect the amount of synthesized signal during each PCR cycle. The relative amounts of the mRNAs were determined using the MxPro program (Stratagene). The amount of PCR product was normalized as a percentage of the expression level of β-actin or GAPDH. The PCR products of OCT4, ZSCAN10, GSS and β-actin were also evaluated on 1.2% agarose gels after staining with ethidium bromide. The sequences of the primers used are provided in Supplementary Table 4.
We confirmed transgene silencing (OCT4, SOX2, KLF4 and MYC) using primers spanning the 5′ region of the viral vector and the 5′ end of the structural genes. Uninfected fibroblasts were used as a negative control and day 3 fibroblasts transfected with Yamanaka factors were used as a positive control. The primer sequences to detect the transgene flanking the pMX vector and the transgene are provided in Supplementary Table 4.
Drug treatments and irradiation
Phleomycin (Sigma) was added at 30 μg ml−1 for 2 h. Cells were processed for analysis 30 min after phleomycin treatment unless indicated otherwise. After a 30 min recovery in ESC medium, the cells were collected and processed for the following experiments. For detection of the DNA damage response in the extended period, the cells were given 6 h to recover after phleomycin treatment and were processed for H2AX immunostaining. In the DNA fragmentation assay, the cells were given 15 h to recover. To check the mutagenesis potential, the cells were treated with 30 μg ml−1 phleomycin for 2 h and cultured for one passage after each treatment. Cells were irradiated at 10 Gy, allowed to recover for 2 h, and then lysates were collected for immunoblot analysis. For the H2O2 treatment (TBHP; stable chemical form of H2O2) cells were treated with 350 μM for 30 min in PBS.
Immunohistochemistry staining
Immunofluorescence analyses were carried out as previously described19. Briefly, cells were fixed in 3.7% formaldehyde for 20 min at room temperature and washed with PBS. Samples were then permeabilized with 0.1 Triton X-100 in PBS for 20 min and blocked for 1 h with 3% BSA in PBS-T, and primary antibodies were incubated for 2 h at room temperature or overnight at 4 °C. Antibody information is provided in Supplementary Table 3. The nuclei were stained with DAPI (Sigma). Alkaline phosphatase staining was carried out using an alkaline phosphatase detection kit (Millipore) according to the manufacturer’s instructions.
Flow cytometry
For the flow cytometry analysis, undifferentiated iPSCs were dissociated with Accutase and stained with mouse anti-human TRA-1-81, anti-human/mouse SSEA-3, and anti-SSEA-4 human/mouse SSEA-4 antibodies. Antibody information is provided in Supplementary Table 3.
HPRT assay
The HPRT gene is on the X chromosome in mammalian cells, and it is used as a model gene to investigate gene mutations in mammalian cell lines32,52. After extended tissue culture of ESCs, Y-iPSCs, A-iPSCs and A-iPSCs–ZSCAN10 with three rounds of phleomycin treatment, 106 ESCs and iPSCs were plated onto 10 cm tissue culture dishes containing feeder cells (CF-1 MEFs) and 5 μg ml−1 of 6-TG (2-amino-6-mercaptopurine, Sigma) was added for negative selection, as only the cells containing the HPRT mutation can grow in 6-TG medium. The mutation frequency was estimated by the inactivation of HPRT promoter activity. Individual colonies were counted/picked on day 12, and the number of colonies was normalized as a percentage of colonies that did not express HPRT in each group by qPCR analysis.
DNA fragmentation analysis
DNA fragmentation was measured using an in situ cell death assay kit (Roche) for visualization of DNA strand breaks by labelling the free 3′-OH termini with modified nucleotides (for example, biotin-dUTP, DIG-dUTP, fluorescein-dUTP) in an enzymatic reaction. iPSC cells (1 × 105 cells) were treated with phleomycin (30 μg ml−1) for 2 h. Samples were collected as control or treated for analysis 15 h after phleomycin treatment. Additionally, cells were treated with DNase I recombinant (Roche) (10 min, 3 U ml−1, at 15 °C to 25 °C) to induce DNA strand breaks, as a positive control for apoptosis. Medium containing floating cells and attached cells was centrifuged (1,000 g, 5 min) and collected. Cells were processed for fluorescence microscopy.
Immunoblot analysis
Treated and untreated cells (1 × 105 cells) were collected 30 min after the 2 h phleomycin treatment (30 μg ml−1). To harvest protein, 100–200 ml RIPA buffer (50 mM Tris-HCl (pH 7.4), 150 mM NaCl, 1% NP40, 0.25% Na-deoxycholate, 1 mM PMSF, protease inhibitor cocktail and phosphatase inhibitor cocktail) was added to floating cell pellets and the remaining adherent cells. The samples were incubated on ice (10 min) and centrifuged (14,000g, 10 min, 4 °C). Protein concentrations were determined using a BCA protein assay kit (Pierce). Samples were adjusted to the same concentration with RIPA buffer (3,000 μg ml−1) and were combined with Laemmli sample buffer (Bio-Rad) and β-mercaptoethanol (Sigma) then heated at 95 °C for 5 min and loaded onto a 4–15% Mini Protean TGX SDS–PAGE gel (Bio-Rad). Samples on the SDS–PAGE gel were transferred to a 0.2 mm polyvinylidene difluoride membrane at 100 V for 1 h, using a wet electro-transfer method (0.2 M glycine, 25 mM Tris and 20% methanol). The membrane was blocked with 5% BSA in PBS-T (1 h at 4 °C), followed by incubation with primary antibodies in blocking solution (5% BSA in PBS containing Tween-20 (1:1,000) (PBS-T), overnight at 4 °C). After primary antibody incubation, membranes were washed three times in PBS-T prior to addition of secondary antibody labelled with peroxidase. Antibody information is provided in Supplementary Table 3. ATM/H2AX negative ESC controls were imported from Xie’s laboratory53 and the ATM negative human fibroblast control was purchased from the Coriell Institute (catalogue no AT5BI, GM05823) (ref. 54).
ChIP–qPCR
ChIP was carried out according to the published protocol55. Immunoprecipitation was carried out with mouse and human ESC and iPSC extracts with IgG or anti-ZSCAN10 (anti-Znf206 antibody, Abcam ab166662) for mouse samples and anti-ZSCAN10 from Novus Biologicals (AF4020) for human samples. We used qPCR to analyse the GSS DNA fragments in the immunoprecipitated samples. Results are presented as ‘percentage input’ values, calculated by using real-time qPCR to quantify the abundance of the DNA fragment of interest added to the ChIP reaction, with respect to the abundance of the DNA fragment found in the final immunoprecipitate. The sequences of the primers used are provided in Supplementary Table 4. The ZSCAN10 binding site on the GSS promoter was estimated based on published ChIP-on-chip data27 and the consensus sequence of ZSCAN10 (5′-[GA]CGCNNGCG[CT]-3′), both of which are highly conserved in mouse and human.
Teratoma analysis of mouse and human iPSCs
Mouse teratomas were assessed by injecting 106 undifferentiated cells into the subcutaneous tissue above the rear haunch of Rag2/γC immunodeficient mice (Taconic), and teratoma formation was monitored for 3 months post-injection. Collected tumours were fixed in 10% formalin solution and processed for haematoxylin and eosin staining by the Molecular Cytology facility of Memorial Sloan Kettering Cancer Center and by Histowiz (http://histowiz.com). For teratoma analysis of human iPSCs, the process was similar; that is, teratomas were assessed by injecting intramuscularly 106 undifferentiated cells along with Matrigel. The source of the cell lines and the methods used for identification and authentication are summarized in Supplementary Table 1. The lines have been tested for mycoplasma contamination.
Cytogenetic analysis and copy number profiling analysis
Cytogenetic analysis was carried out by metaphase chromosome preparation and G-band karyotyping at the Molecular Cytogenetics Core Facility of Memorial Sloan Kettering Cancer Center. Copy number profiling analysis was carried out according to the published protocol43. The gains of chromosomes 12 and 17 were disregarded as common trisomies observed in human ESCs56–59.
Growth rate
10,000 cells per line were plated in triplicate in 24-well dishes, and the numbers of cells were counted every 24 h for 3 days.
H2O2 ROS assay (DCFDA assay)
H2O2 scavenging activity was measured using a cellular ROS assay kit (Abcam, ab113851). ESCs/iPSCs were labelled with 20 μM DCFDA (2′,7′-dichlorofluorescein diacetate; a fluorogenic dye that measures hydroxyl, peroxyl and other ROS activity within the cell), and cultured for 3 h with 50 μM TBHP (stable chemical form of H2O2). Cells were then analysed on a fluorescent plate reader. Mean ± s.d. is plotted for four replicates from each condition.
TBHP treatment
Cells were treated with 350 μM TBHP solution (Luperox TBH70X, TBHP solution 70 wt% in H2O, 458139) for 30 min in PBS. Lysates were collected for immunoblot analysis. The control untreated cell lines were cultured in either ESC medium or PBS without TBHP treatment, and DNA damage response was not induced (data not shown).
Glutathione detection assay
Feeder-free cells were cultured on Matrigel-coated tissue culture plates in MEF-conditioned ESC medium. On day 3, the cells were washed in PBS and scraped and pelleted by centrifugation. Subsequent steps were carried out using a glutathione fluorometric assay kit (catalogue no K264-100, BioVision) according to the manufacturer’s manual. Briefly, cell pellets were homogenized in ice cold glutathione assay buffer, preserved in perchloric acid and centrifuged. Supernatants were neutralized with potassium hydroxide. After centrifugation, either the supernatant was used to detect reduced glutathione (GSH), or total glutathione was measured by reducing oxidized glutathione (GSSG) to GSH before measurement. For measuring GSSG concentrations specifically, existing GSH was quenched before reducing agent was applied. o-phthalaldehyde probe, which reacts with GSH and emits fluorescence, was added to samples, and signal was acquired at Ex/Em = 340 nm/420 nm on a Varioskan Flash by Thermo Scientific. The oxidation capacity of glutathione was determined by the quantity of total glutathione (GSH+GSSG).
Generation of A-iPSCs–shGSS and Y-iPSCs–GSS
A-iPSCs were infected post-reprogramming with a set of shRNA viruses for GSS. For the generation of A-iPSCs–shGSS, A-iPSCs were infected post-reprogramming with a set of shRNA viral vectors for GSS (GE Dharmacon, RMM4532-EG14854 and GE Dharmacon RHS453-EG2937 for mouse and human respectively). Clones were selected with puromycin, and the levels of downregulation were measured by qPCR. For the generation of Y-iPSCs–GSS, Y-iPSCs were infected with a lentivirus carrying the GSS cDNA (Harvard Plasmid Core (http://plasmid.med.harvard.edu/PLASMID) post-reprogramming. The infected clones were sorted for a red fluorescent marker and the GSS expression levels were assessed by qPCR.
Gene expression analysis
Illumina genome-wide gene expression arrays (MouseRef-8 v2.0 MouseRef-8 v2.0 BeadChip) were used for mRNA expression analysis of Y-somatic cells (Y-SC fibroblasts), A-somatic cells (A-SC fibroblasts), ESCs, Y-iPSCs, A-iPSCs and A-iPSCs–ZSCAN10.
Sample and exome library preparation
Genomic DNA samples from patient-derived cell lines (hiPSCs) were obtained using a DNeasy blood and tissue kit (Qiagen), and whole exome sequencing was carried out at the Mayo Clinic Advanced Genomics Technology Center. Paired-end libraries using 1.0 μg of gDNA were prepared according to the standard manufacturer’s instructions with the Agilent Bravo liquid handler. The concentration and size distribution of the completed libraries was determined using an Agilent DNA 1000 bioanalyser chip and Qubit fluorometry (Invitrogen). Whole exome capture was carried out using 750 ng of the prepped library following the protocol for the SureSelect Human All Exon v5 + UTRs 75 MB kit (Agilent). The purified capture products were then amplified using the SureSelect Post-Capture Indexing forward and Index PCR reverse primers (Agilent) for 12 cycles. Libraries were sequenced at an average coverage of 80× following Illumina’s standard protocol using the Illumina cBot and cBot paired-end cluster kit v3. The flow cells were sequenced as 101 × 2 paired-end reads on an Illumina HiSeq 2000 using a TruSeq SBS sequencing kit v3 and HCS v2.0.12 data collection software. Base-calling was carried out using Illumina’s RTA v1.17.21.3 (ref. 60).
Computational analysis of whole exome sequencing
Substitutions and indels were called using a consensus calling approach that included SpeedSeq v0.1.0 (ref. 61) with default parameters. In brief, alignment was conducted with bwa-mem v0.7.12 against UCSC hg19 reference build, deduplicated using samblaster v0.1.22, and somatic calling done using freebayes v0.9.21 using the SpeedSeq parameters (pooled-discrete–genotype-qualities–min-alternate-fraction 0.05–min-alternate-count 2–min-repeat-entropy 1). Variant calling across samples was carried out for exonic regions with minimum sequencing depth of 20 (DP ≥ 20) using bedtools v2.2.21, resulting in covering 60,301,486 bases. A consensus quality score of at least 20 (Q≥ 20) and allele frequency of 30 (AF≥ 0.3) was achieved with VCFtools. A matching aged fibroblast cell line (AG4) was used for germline variant calling in A-hiPSC and A-hiPSC–ZCAN10 cell lines. Variants that occurred at locations present in dbSNP (build137), BGI62 or ESP6500 (http://evs.gs.washington.edu/EVS), 1,000 genome63,64 databases were removed from consideration. Annotation variant analysis was carried out with the BioR variant annotation platform65 against Ensembl v.70 and consequence prediction using SNPeffect (v.2.0.5d) and VEP (v.2.7).
Chimaera
Chimaera analysis of pluripotent cells was conducted by injecting GFP+ into blastocysts isolated from C57BL/6 (GFP–) embryos. The reconstituted blastocysts were implanted into 2.5-day-pseudopregnant CD1 females. These procedures were carried out at the MSKCC transgenic core facility. Chimaeras were collected at embryonic day 12.5 for imaging. The animal study is compliant with all relevant ethical regulations regarding animal research per the Institutional Animal Care and Use Committee (IACUC approval number 11-10-023).
Statistics and reproducibility
Data were analysed using GraphPad Prism 5.0.1 software (GraphPad Software) and/or Excel software (Microsoft Office). Data are presented as mean with s.e.m. or s.d. (as indicated in the figure legends). Statistical tests were carried out and P value thresholds were obtained using GraphPad 5.0.1. or Excel. Multiple groups were tested using two-sided t-test followed by post hoc Holm–Bonferroni correction for a significance level of 0.05 and comparisons between two groups were carried out using two-tailed unpaired Student’s t-test. To confirm reproducibility, every experiment was repeated independently. Representative figures are shown in Figs 1b and 2a and Supplementary Figs 1a,b,d,g,h, 3a,b,d,e, 5a,b,h,i and 6a,b. For representative Figs 1b and 2a, the exact number of independent clones is defined in the quantification experiments in Figs 1c and 2b, respectively, and detailed data are provided in Supplementary Table 5. For representative Supplementary Figs 1a,b,d and 5a,b, detailed information regarding repetitions is provided in Supplementary Table 1. For representative Supplementary Fig. 1g,h, detailed information regarding repetitions is provided in Fig. 1a and in Supplementary Table 5. For representative Supplementary Figs 3a,b, 5i and 6a,b, the experiments were repeated twice. For representative Supplementary Fig. 3d, detailed information can be found in Fig. 2c; for representative Supplementary Fig. 3e, details are found in Supplementary Table 5. For representative Supplementary Fig. 5h, details of the number of independent repeats are indicated in Fig. 6e–g.
Code availability
All the codes used in this study are published.
Data availability
Deep-sequencing and microarray data that support the findings of this study have been deposited in the Gene Expression Omnibus (GEO) under accession codes SRP081214 and GSE85365, respectively. Source data for Figs 1a,c, 2b and 3c,e,f, and Supplementary Figs 1i,j, 3f,h, 4b,c and 5g, are provided in Supplementary Table 5. Unprocessed immunoblots have been provided for Figs 1d,e, 2c, 4a,b, 5a,d and 6c,d and Supplementary Fig. 1i,j,f in Supplementary Fig. 7. All other data supporting the findings of this study are available from the corresponding author on request.
Supplementary Material
Acknowledgments
K.K. is supported by the NIH (R00HL093212), NIH (R01AG043531), TriStem-Star Foundation (2013-049), Louis V. Gerstner, Jr. Young Investigators awards, Geoffrey Beene Junior Chair Award, Sidney Kimmel Scholar Award, Alfred W. Bressler Scholars Endowment Fund and MSKCC Society Fund. MSKCC Core Facilities are supported by NIH Cancer Center support grant P30 CA008748. R.Z. is supported by the UAB Development Fund. H.L. is supported by the NIH (CA196631-01A1) and Mayo Clinic Center for Individualized Medicine. C.-W.L. is supported by the NIH (R21HD081682). J.J.C. is supported by the HHMI and NIH (R24DK092760). S.L. is supported by the HHMI and NIH (P01CA087497). J.K. is supported by the NIH (R01GM112722). H.F. is supported by the TriStem-Star Foundation (2013-049). J.A. acknowledges support from the medical faculty of Heinrich Heine University, Düsseldorf. T.B. is supported by the MSKCC Single Cell Sequencing Initiative, William and Joyce O’Neil Research Fund and STARR visiting fellows programme. S.G. is supported by the American Italian Cancer Foundation and a Women in Science Rockefeller fellowship.
Footnotes
Note: Supplementary Information is available in the online version of the paper
AUTHOR CONTRIBUTIONS
M.S., R.Z., and K.K. conceived the experimental plan. M.S., P.Y., T.B., L.D., J.W., S.G., Z.L., M.B., R.Z. and K.K. carried out the experiments. C.C., T.B., S.B., C.Z., C.A.R., J.K. and H.L. carried out computational analysis. M.S., C.C., P.Y., T.B., C.A.R., S.B., J.A., H.F., J.K., S.L., J.J.C., C.-W.L., H.L., R.Z. and K.K. wrote the manuscript.
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
References
- 1.Prigione A, et al. Mitochondrial-associated cell death mechanisms are reset to an embryonic-like state in aged donor-derived iPS cells harboring chromosomal aberrations. PLoS ONE. 2011;6:e27352. doi: 10.1371/journal.pone.0027352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kang E, et al. Age-related accumulation of somatic mitochondrial DNA mutations in adult-derived human iPSCs. Cell Stem Cell. 2016;18:625–636. doi: 10.1016/j.stem.2016.02.005. [DOI] [PubMed] [Google Scholar]
- 3.Lo Sardo V, et al. Influence of donor age on induced pluripotent stem cells. Nat Biotechnol. 2016;35:69–74. doi: 10.1038/nbt.3749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Garber K. RIKEN suspends first clinical trial involving induced pluripotent stem cells. Nat Biotechnol. 2015;33:890–891. doi: 10.1038/nbt0915-890. [DOI] [PubMed] [Google Scholar]
- 5.Coghlan A. Unexpected mutations put stem cell trial on hold. New Sci. 2015;227:9. [Google Scholar]
- 6.Kang E, et al. Mitochondrial replacement in human oocytes carrying pathogenic mitochondrial DNA mutations. Nature. 2016;540:270–275. doi: 10.1038/nature20592. [DOI] [PubMed] [Google Scholar]
- 7.Mandai M, et al. Autologous induced stem-cell-derived retinal cells for macular degeneration. New Engl J Med. 2017;376:1038–1046. doi: 10.1056/NEJMoa1608368. [DOI] [PubMed] [Google Scholar]
- 8.Holmstrom KM, Finkel T. Cellular mechanisms and physiological consequences of redox-dependent signalling. Nat Rev Mol Cell Biol. 2014;15:411–421. doi: 10.1038/nrm3801. [DOI] [PubMed] [Google Scholar]
- 9.Bigarella CL, Liang R, Ghaffari S. Stem cells and the impact of ROS signaling. Development. 2014;141:4206–4218. doi: 10.1242/dev.107086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liochev SI. Reactive oxygen species and the free radical theory of aging. Free Radic Biol Med. 2013;60:1–4. doi: 10.1016/j.freeradbiomed.2013.02.011. [DOI] [PubMed] [Google Scholar]
- 11.Baccelli I, Trumpp A. The evolving concept of cancer and metastasis stem cells. J Cell Biol. 2012;198:281–293. doi: 10.1083/jcb.201202014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Alfadda AA, Sallam RM. Reactive oxygen species in health and disease. J Biomed Biotechnol. 2012;2012:936486. doi: 10.1155/2012/936486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sleigh MJ. The mechanism of DNA breakage by phleomycin in vitro. Nucl Acids Res. 1976;3:891–901. doi: 10.1093/nar/3.4.891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Franco R, Cidlowski JA. Apoptosis and glutathione: beyond an antioxidant. Cell Death Differ. 2009;16:1303–1314. doi: 10.1038/cdd.2009.107. [DOI] [PubMed] [Google Scholar]
- 15.Harris IS, et al. Glutathione and thioredoxin antioxidant pathways synergize to drive cancer initiation and progression. Cancer Cell. 2015;27:211–222. doi: 10.1016/j.ccell.2014.11.019. [DOI] [PubMed] [Google Scholar]
- 16.Guo Z, Kozlov S, Lavin MF, Person MD, Paull TT. ATM activation by oxidative stress. Science. 2010;330:517–521. doi: 10.1126/science.1192912. [DOI] [PubMed] [Google Scholar]
- 17.Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- 18.Smith KP, Luong MX, Stein GS. Pluripotency: toward a gold standard for human ES and iPS cells. J Cell Physiol. 2009;220:21–29. doi: 10.1002/jcp.21681. [DOI] [PubMed] [Google Scholar]
- 19.Kim K, et al. Epigenetic memory in induced pluripotent stem cells. Nature. 2010;467:285–290. doi: 10.1038/nature09342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu JC, Lerou PH, Lahav G. Stem cells: balancing resistance and sensitivity to DNA damage. Trends Cell Biol. 2014;24:268–274. doi: 10.1016/j.tcb.2014.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu JC, et al. High mitochondrial priming sensitizes hESCs to DNA-damage-induced apoptosis. Cell Stem Cell. 2013;13:483–491. doi: 10.1016/j.stem.2013.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhivotovsky B, Kroemer G. Apoptosis and genomic instability. Nat Rev Mol Cell Biol. 2004;5:752–762. doi: 10.1038/nrm1443. [DOI] [PubMed] [Google Scholar]
- 23.Gurdon JB, Wilmut I. Nuclear transfer to eggs and oocytes. Cold Spring Harb Perspect Biol. 2011;3:1–14. doi: 10.1101/cshperspect.a002659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ma H, et al. Abnormalities in human pluripotent cells due to reprogramming mechanisms. Nature. 2014;511:177–183. doi: 10.1038/nature13551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cowan CA, Atienza J, Melton DA, Eggan K. Nuclear reprogramming of somatic cells after fusion with human embryonic stem cells. Science. 2005;309:1369–1373. doi: 10.1126/science.1116447. [DOI] [PubMed] [Google Scholar]
- 26.Kim J, Chu J, Shen X, Wang J, Orkin SH. An extended transcriptional network for pluripotency of embryonic stem cells. Cell. 2008;132:1049–1061. doi: 10.1016/j.cell.2008.02.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yu HB, Kunarso G, Hong FH, Stanton LW. Zfp206, Oct4, and Sox2 are integrated components of a transcriptional regulatory network in embryonic stem cells. J Biol Chem. 2009;284:31327–31335. doi: 10.1074/jbc.M109.016162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang W, et al. Zfp206 regulates ES cell gene expression and differentiation. Nucl Acids Res. 2006;34:4780–4790. doi: 10.1093/nar/gkl631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ford J, Ahmed S, Allison S, Jiang M, Milner J. JNK2-dependent regulation of SIRT1 protein stability. Cell Cycle. 2008;7:3091–3097. doi: 10.4161/cc.7.19.6799. [DOI] [PubMed] [Google Scholar]
- 30.Jiang J, et al. Zscan4 promotes genomic stability during reprogramming and dramatically improves the quality of iPS cells as demonstrated by tetraploid complementation. Cell Res. 2013;23:92–106. doi: 10.1038/cr.2012.157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chan EM, et al. Live cell imaging distinguishes bona fide human iPS cells from partially reprogrammed cells. Nat Biotechnol. 2009;27:1033–1037. doi: 10.1038/nbt.1580. [DOI] [PubMed] [Google Scholar]
- 32.Tsuda H, Sasaki K, Tanaka N. Establishment of hypoxanthine phosphoribosyltransferase(HPRT)-locus mutation assay system in mouse ES cells. Altern Anim Test Exp. 2005;11:118–128. [Google Scholar]
- 33.Balendiran GK, Dabur R, Fraser D. The role of glutathione in cancer. Cell Biochem Funct. 2004;22:343–352. doi: 10.1002/cbf.1149. [DOI] [PubMed] [Google Scholar]
- 34.Estrela JM, Ortega A, Obrador E. Glutathione in cancer biology and therapy. Crit Rev Clin Lab Sci. 2006;43:143–181. doi: 10.1080/10408360500523878. [DOI] [PubMed] [Google Scholar]
- 35.Traverso N, et al. Role of glutathione in cancer progression and chemoresistance. Oxid Med Cell Longev. 2013;2013:972913. doi: 10.1155/2013/972913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Miller JD, et al. Human iPSC-based modeling of late-onset disease via progerin-induced aging. Cell Stem Cell. 2013;13:691–705. doi: 10.1016/j.stem.2013.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kyttala A, et al. Genetic variability overrides the impact of parental cell type and determines iPSC differentiation potential. Stem Cell Rep. 2016;6:200–212. doi: 10.1016/j.stemcr.2015.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Takahashi K, et al. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131:861–872. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- 39.Nakagawa M, et al. Generation of induced pluripotent stem cells without Myc from mouse and human fibroblasts. Nat Biotechnol. 2008;26:101–106. doi: 10.1038/nbt1374. [DOI] [PubMed] [Google Scholar]
- 40.Araki R, et al. Crucial role of c-Myc in the generation of induced pluripotent stem cells. Stem Cells. 2011;29:1362–1370. doi: 10.1002/stem.685. [DOI] [PubMed] [Google Scholar]
- 41.Okita K, et al. A more efficient method to generate integration-free human iPS cells. Nat Methods. 2011;8:409–412. doi: 10.1038/nmeth.1591. [DOI] [PubMed] [Google Scholar]
- 42.Unternaehrer JJ, et al. The epithelial-mesenchymal transition factor SNAIL paradoxically enhances reprogramming. Stem Cell Rep. 2014;3:691–698. doi: 10.1016/j.stemcr.2014.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Baslan T, et al. Optimizing sparse sequencing of single cells for highly multiplex copy number profiling. Genome Res. 2015;25:714–724. doi: 10.1101/gr.188060.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yamamoto S, et al. Rapid selection of XO embryonic stem cells using Y chromosome-linked GFP transgenic mice. Transgenic Res. 2014;23:757–765. doi: 10.1007/s11248-014-9813-0. [DOI] [PubMed] [Google Scholar]
- 45.Seol HW, et al. Separation and maintenance of normal cells from human embryonic stem cells with trisomy 12 mosaicism. Chromosome Res. 2008;16:1075–1084. doi: 10.1007/s10577-008-1258-y. [DOI] [PubMed] [Google Scholar]
- 46.Kilpinen H, et al. Common genetic variation drives molecular heterogeneity in human iPSCs. Nature. 2017;546:370–375. doi: 10.1038/nature22403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Finkel T, Holbrook NJ. Oxidants, oxidative stress and the biology of ageing. Nature. 2000;408:239–247. doi: 10.1038/35041687. [DOI] [PubMed] [Google Scholar]
- 48.Shaw PX, Werstuck G, Chen Y. Oxidative stress and aging diseases. Oxid Med Cell Longev. 2014;2014:569146. doi: 10.1155/2014/569146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Son J, et al. Glutamine supports pancreatic cancer growth through a KRAS-regulated metabolic pathway. Nature. 2013;496:101–105. doi: 10.1038/nature12040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hollander M, Wolfe DA. Nonparametric Statistical Methods. Wiley; 1973. [Google Scholar]
- 51.Kim K, et al. Donor cell type can influence the epigenome and differentiation potential of human induced pluripotent stem cells. Nat Biotechnol. 2011;29:1117–1119. doi: 10.1038/nbt.2052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Johnson GE. Mammalian cell HPRT gene mutation assay: test methods. Methods Mol Biol. 2012;817:55–67. doi: 10.1007/978-1-61779-421-6_4. [DOI] [PubMed] [Google Scholar]
- 53.Rass E, Chandramouly G, Zha S, Alt FW, Xie A. Ataxia telangiectasia mutated (ATM) is dispensable for endonuclease I-SceI-induced homologous recombination in mouse embryonic stem cells. J Biol Chem. 2013;288:7086–7095. doi: 10.1074/jbc.M112.445825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hahn P, Kapp LN, Painter RB. Establishment and characterization of two human cell lines with amplified dihydrofolate reductase genes. Exp Cell Res. 1987;168:89–94. doi: 10.1016/0014-4827(87)90418-6. [DOI] [PubMed] [Google Scholar]
- 55.Tian B, Yang J, Brasier AR. Two-step cross-linking for analysis of protein–chromatin interactions. Methods Mol Biol. 2012;809:105–120. doi: 10.1007/978-1-61779-376-9_7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Buzzard JJ, Gough NM, Crook JM, Colman A. Karyotype of human ES cells during extended culture. Nat Biotechnol. 2004;22:381–382. doi: 10.1038/nbt0404-381. [DOI] [PubMed] [Google Scholar]
- 57.Imreh MP, et al. In vitro culture conditions favoring selection of chromosomal abnormalities in human ES cells. J Cell Biochem. 2006;99:508–516. doi: 10.1002/jcb.20897. [DOI] [PubMed] [Google Scholar]
- 58.Li SS, et al. Characterization and gene expression profiling of five new human embryonic stem cell lines derived in Taiwan. Stem Cells Dev. 2006;15:532–555. doi: 10.1089/scd.2006.15.532. [DOI] [PubMed] [Google Scholar]
- 59.Plaia TW, et al. Characterization of a new NIH-registered variant human embryonic stem cell line, BG01V: a tool for human embryonic stem cell research. Stem Cells. 2006;24:531–546. doi: 10.1634/stemcells.2005-0315. [DOI] [PubMed] [Google Scholar]
- 60.Altmann HM, et al. Homozygous/compound heterozygous triadin mutations associated with autosomal-recessive long-QT syndrome and pediatric sudden cardiac arrest: elucidation of the triadin knockout syndrome. Circulation. 2015;131:2051–2060. doi: 10.1161/CIRCULATIONAHA.115.015397. [DOI] [PubMed] [Google Scholar]
- 61.Chiang C, et al. SpeedSeq: ultra-fast personal genome analysis and interpretation. Nat Methods. 2015;12:966–968. doi: 10.1038/nmeth.3505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Li Y, et al. Resequencing of 200 human exomes identifies an excess of low-frequency non-synonymous coding variants. Nat Genet. 2010;42:969–972. doi: 10.1038/ng.680. [DOI] [PubMed] [Google Scholar]
- 63.The 1000 Genomes Project Consortium. A map of human genome variation from population-scale sequencing. Nature. 2010;467:1061–1073. doi: 10.1038/nature09534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.The 1000 Genomes Project Consortium. An integrated map of genetic variation from 1,092 human genomes. Nature. 2012;491:56–65. doi: 10.1038/nature11632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kocher JP, et al. The Biological Reference Repository (BioR): a rapid and flexible system for genomics annotation. Bioinformatics. 2014;30:1920–1922. doi: 10.1093/bioinformatics/btu137. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Deep-sequencing and microarray data that support the findings of this study have been deposited in the Gene Expression Omnibus (GEO) under accession codes SRP081214 and GSE85365, respectively. Source data for Figs 1a,c, 2b and 3c,e,f, and Supplementary Figs 1i,j, 3f,h, 4b,c and 5g, are provided in Supplementary Table 5. Unprocessed immunoblots have been provided for Figs 1d,e, 2c, 4a,b, 5a,d and 6c,d and Supplementary Fig. 1i,j,f in Supplementary Fig. 7. All other data supporting the findings of this study are available from the corresponding author on request.