Extended Data Figure 1. Thermo-stability of DRD2 constructs, crystal packing of the DRD2/Risperidone complex and representative electron density of the DRD2 structure.
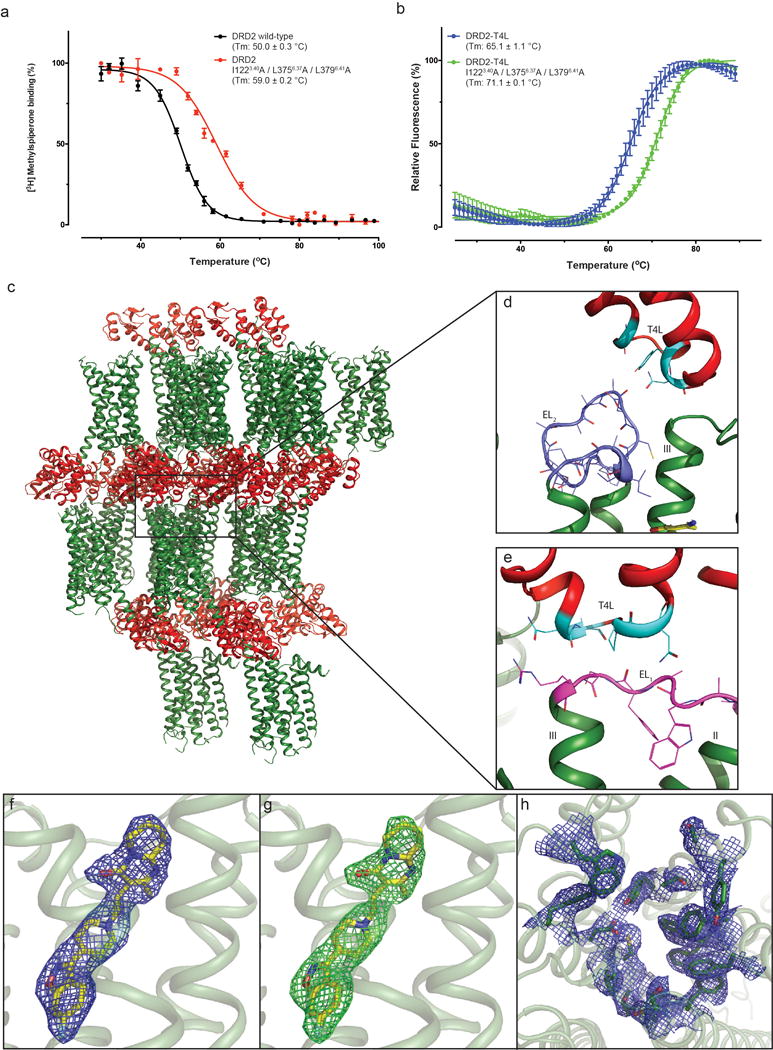
a, DRD2 or thermo-stability mutation membrane with 1nM [3H]-N-methylspiperone were heated for 30 min, the amount of [3H]-ligand bound determined. b, Purified DRD2-T4L (with or without thermo-stability mutation) protein with 10 μM risperidone and 1 μM BODIPY FL L-cystine dye were heated by a temperature gradient and the amount of dye bound to unfolding protein determined. Data were analyzed by nonlinear regression and apparent Tm values (transition temperature where 50% of the receptor is inactive) were determined from analysis of the sigmoidal dose-response curves. All data in a-b are the mean SEM of three independent assays. Error bars in a-b denote SEM from three independent assays. c, d, e, Packing of the DRD2/Risperidone complex crystallized in the P212121 spacegroup. The DRD2 is shown in green and T4L fusion protein is shown in red or cyan (interact with DRD2). EL1 and EL2 of DRD2 were shown in magenta and blue, respectively. f, 2Fo-Fc electron density map (blue mesh) of risperidone (yellow) contoured at 1σ. g, Fo-Fc omit map (green mesh) contoured at 3.0σ of risperidone (yellow). h, 2Fo-Fc electron density map of DRD2 binding pocket residues (blue mesh) contoured at 1σ.
