Extended Data Figure 2. Conserved hydrophobic residue of EL2 in all available aminergic receptor structures.
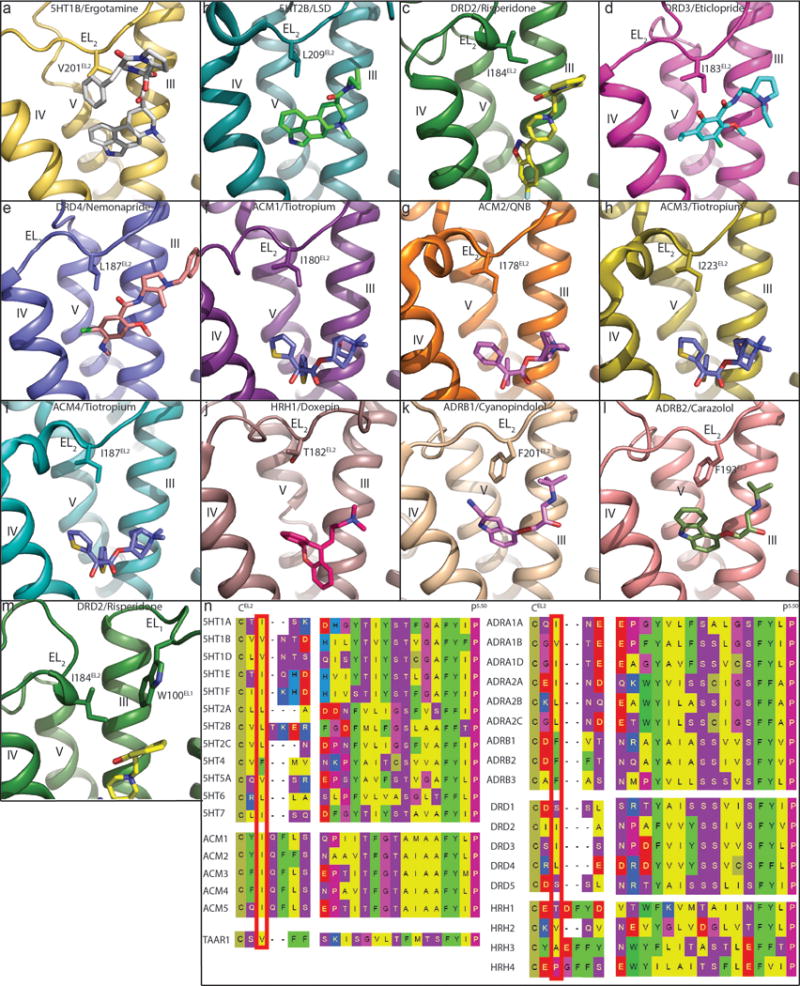
In all panels, receptors are shown as cartoon. Ligands and residues are shown as sticks. a, 5HT1B (PDB code 4IAR). b, 5HT2B (PDB code 5TVN). c, DRD2. d, DRD3 (PDB code 3PBL). e, DRD4 (PDB code 5WIU). f, ACM1 (PDB code 5CXV). g, ACM2 (PDB code 3UON). h, ACM3 (PDB code 4ADJ). i, ACM4 (PDB code 4DSG). j, HRH1 (PDB code 3RZE). k, ADRB1 (PDB code 2VT4). l, ADRB2 (PDB code 2RH1). m, DRD2. n, Conserved EL2 hydrophobic residues (red box) are located two residues away from conserved cysteine that forms a disulfide bridge between EL2 and helix III. Notable exceptions to the presence of a hydrophobic residue are DRD1 and DRD5, which contain a serine, and HRH1 and HRH4, which contain a threonine and proline, respectively.
