Abstract
Cell adhesion in the multiple myeloma (MM) microenvironment has been recognized as a major mechanism of MM cell survival and the development of drug resistance. Here we addressed the hypothesis that the protein junctional adhesion molecule-A (JAM-A) may represent a novel target and a clinical biomarker in MM. We evaluated JAM-A expression in MM cell lines and in 147 MM patient bone marrow aspirates and biopsies at different disease stages. Elevated JAM-A levels in patient-derived plasma cells were correlated with poor prognosis. Moreover, circulating soluble JAM-A (sJAM-A) levels were significantly increased in MM patients as compared with controls. Notably, in vitro JAM-A inhibition impaired MM migration, colony formation, chemotaxis, proliferation and viability. In vivo treatment with an anti-JAM-A monoclonal antibody (αJAM-A moAb) impaired tumor progression in a murine xenograft MM model. These results demonstrate that therapeutic targeting of JAM-A has the potential to prevent MM progression, and lead us to propose JAM-A as a biomarker in MM, and sJAM-A as a serum-based marker for clinical stratification.
Introduction
Multiple myeloma (MM) is caused by the clonal expansion of malignant MM plasma cells (MM-PCs) in the bone marrow (BM). MM remains incurable, despite advances in conventional therapy, clinical application of novel agents and high-dose chemotherapy protocols supported by autologous stem cell transplantation.1 Biological stratification2 and drug resistance are major challenges in treating MM.3 The tumor microenvironment appears to have a pivotal role in disease progression. Interactions between MM-PCs and BM stromal cells (BMSCs) promote refractory disease and are correlated with tumor growth and relapse,4 involving several molecules.5 MM cells in contact with BMSCs and extracellular matrix components escape the effects of therapy through cell adhesion-mediated drug resistance.6, 7 MM-PCs achieve this by means of molecules of the integrin family, CD44, Syndecan-1 (CD138), Lymphocyte Function-Associated Antigen-1 (LFA-1), Mucin-1 antigen (MUC-1), Vascular Cell Adhesion Molecule-1 (VCAM-1) and Intercellular Adhesion Molecule-1 (ICAM-1).8, 9, 10, 11 In line with several observations,12, 13, 14, 15 Paiva et al.16 recently demonstrated that integrins and adhesion molecules are overexpressed in MM cells from patients with minimal residual disease compared with MM-PCs from newly diagnosed MM patients. MM-PC-BMSC adhesion triggers interleukin-6 secretion, NF-κB activation in BMSCs17 as well as the upregulation of signaling pathways in MM cells allowing cell proliferation and survival.18 Hence, the adhesion/migration systems are key targets19 in MM treatment. Herein, we investigated contact-induced receptors upon physical interaction of MM and bone cells.20 We identified members of the junctional adhesion molecule (JAM) family and the chemokine receptor CXCR4 as contact-enhanced targets: first,21 CXCR4 has been described as a useful theranostic target in MM patients;22 second, JAM-A is associated with CXCR4;23 and third, JAM-A was described to have an important role in hematopoietic stem cell biology.24 These observations prompted us to investigate JAM-A in MM. JAM-A is a type I transmembrane glycoprotein belonging to the immunoglobulin superfamily.25 JAM-A is widely expressed in human tissues,26 predominantly in intercellular junctions of epithelial and endothelial cells and on the surface of leukocytes, lymphocytes, platelets and erythrocytes.27 JAM-A is implicated in cell–cell adhesion, leukocyte migration, platelet activation, angiogenesis and reovirus binding.28 The JAM-A protein is composed of an extracellular domain with two Ig-like loops, a single membrane-spanning region and a short cytoplasmic tail terminating in a PDZ-binding motif. Homophilic interactions are important for the above-mentioned JAM-A functions in cells.29 The C-terminal PDZ binding motif can facilitate interactions with various scaffold proteins, such as Zona Occludens 1 (ZO-1), Afadin-6 (AF-6) and Partitioning defective 3 homolog (PARD3).27 JAM-A dimerization and PDZ-binding motifs are critical in triggering a functional intracellular signaling cascade.27, 28, 29 JAM-A is a single transmembrane protein with an extracellular domain containing a cleavage site.30, 31 Circulating soluble JAM-A (sJAM-A) is associated with inflammation, angiogenesis and vascular diseases.32 However, the significance of variations of plasma sJAM-A remain unclear.26 Recently, JAM-A has been implicated in solid cancer progression, but even though JAM-A has been associated with invasion, metastasis and poor prognosis in several solid tumors,33, 34, 35, 36, 37, 38 its role in hematological malignancies is still unclear. Immunophenotypic studies of JAM-A expression in patients with MM are scanty26, 39 and its potential biologic and prognostic value remains unknown.39
Here, we show a correlation between high JAM-A expression in MM patients and poor clinical prognosis. Blocking JAM-A in vitro impaired MM cell viability, proliferation, migration and chemotaxis, and significantly reduced MM progression in vivo and dissemination in MM-bearing mice. On the basis of these findings, we propose JAM-A as a promising novel therapeutic target in the treatment of MM.
Materials and methods
Ethics statement
Approval for the study was obtained from the Institutional Ethics Committee of Würzburg University Hospital. Written informed consent was obtained from all MM patients and all clinical investigations were conducted in accordance with the Declaration of Helsinki. BM aspirates were collected from all patients and controls. All animal experiments were performed in accordance with the German regulations for animal experimentation.
MM patient samples
A total of 147 subjects (63% males and 37% females) was included in our study, comprising 73 patients with newly diagnosed MM, 65 patients who had received previous treatments, among whom 49 patients were with optimal response (22 patients either with complete response or 27 patients with very good partial response, respectively) and 16 patients with suboptimal response MM (15 patients either with partial response or 1 with stable disease, respectively). In nine cases complete clinical and laboratory information was not available for all time points of analysis (Supplementary Table 1). All patients were treated between December 2011 and July 2016. Three follow-up visits were scheduled: at the time of recruitment (t0), after receiving at least four cycles of therapy (t1) and at the time of relapse (t2; Supplementary Table 1). Disease stages included stage I (n=61), II (n=53) and III (n=25). Genetic risk was assessed using fluorescent in situ hybridization at the time of recruitment. Accordingly, high genetic risk was defined as the presence of del(17p); t(14;16); t(14;20); intermediate risk as the presence of −13/del(13q) or t(4;14), that constituted high risk only if associated with Beta-2 Microglobulin (B2MG)>4 mg/l and Hb<10 g/dl. Finally, the standard risk group included all cytogenetic abnormalities not mentioned above, including translocations t(11;14); t(6;14).1, 2 Eleven monoclonal gammopathy of undetermined significance (MGUS) patients and 19 healthy donors were included (details in Supplementary Table 2).
Cell lines and cell culture conditions
RPMI-8226, U266, OPM-2 and NCI-H929 cell lines were obtained from the Leibniz Institute (DSMZ-German Collection of Microorganisms and Cell Culture, Braunschweig, Germany). Human umbilical vein endothelial cells (HUVECs) were obtained from PromoCell (Heidelberg, Germany). Interleukin-6-dependent human MM cell line INA-640—as a model for cytokine-regulated plasmocytoma—was a kind gift from M Gramatzki (Kiel, Germany). Details are provided in Supplementary Methods.
Cell preparation for immunofluorescence microscopy
BM mononuclear cells were isolated from heparinized BM aspirates via Ficoll gradient. Malignant plasma cells (MM-PCs) were obtained with magnetic anti-CD138 beads (Immunotech, Vaudreuil-Dorion, Canada), whereas adherent CD138 cells were cultured separately as BMSCs. Culture supernatants were centrifuged (× 200g for 10 min) and stored at −80 °C as conditioned media. Immunofluorescence analyses were performed on BM from MM and MGUS patients and cultured cells, MM-PCs, RPMI-8226 and control PCs, PBMCs and HUVECs. Details are supplied in Supplementary Methods.
Flow cytometry
After immunomagnetic enrichment, MM-PCs, obtained from BM aspirates, were identified as the CD138+ population within the gate of live cells. MM-PCs, MM cell lines, HUVECs and PBMCs were stained with an anti-JAM-A antibody (anti-JAM-A FITC, clone OV5B8 (BioLegend, San Diego, CA, USA)). Samples were acquired with the fluorescence activated cell sorting (FACS) Canto II flow cytometer (BD Biosciences, San Jose, CA, USA).
Immunohistochemical analysis of BM biopsies
We established immunohistochemical staining protocols for JAM-A in BM biopsies to investigate the protein expression levels in situ in human MGUS- and MM-PCs. Details are supplied in Supplementary Methods.
Enzyme-linked immunosorbent assay
Circulating sJAM-A was quantified in the sera from healthy controls and MM patients with an enzyme-linked immunosorbent assay (ELISA) kit for human JAM-A (R&D Systems, Minneapolis, MN, USA), according to the manufacturer’s instructions.
Quantitative real-time PCR
Quantitative real-time PCR was performed using SYBR Green PCR master mix (Applied Biosystems, Foster City, CA, USA) according to the manufacturer’s instructions. Details are provided in Supplementary Methods.
Small interfering RNA treatment
Small interfering RNA (siRNA) oligonucleotides (Sigma-Aldrich, St Louis, MO, USA) against human JAM-A were used as follows: 5′-GAAGUGAAGGAGAAUUCAATT-3′ (sense) and 5′-UUGAAUUCUCCUUCACUUCTT-3′ (antisense). The sequences of non-targeting siRNA oligonucleotides used as a negative control were as follows: 5′-UUCUCCGAACGUGUCACGUTT-3′ (sense) and 5′-ACGUGACACGUUCG GAGAATT-3′ (antisense). Details are supplied in Supplementary Methods.
Scratch assay
Confluent RPMI-8226 on fibronectin (10 μg/ml)-coated (Sigma-Aldrich) 6 cm2 dishes were scraped as a ‘wound’ with a pipette tip. Additional details are provided in Supplementary Methods.
Colony-forming assay
MM cell colonies were evaluated as previously described (details in Supplementary Methods).41
Chemotaxis
Chemotaxis was performed using the ThinCert assay (Greiner Bio-One GmbH, Frickenhausen, Germany). Details are provided in Supplementary Methods. Ki-67 evaluation and apoptosis assays are described in Supplementary Methods.
RPMI-8226 luc+ cell line and culture conditions
RPMI-8226 cells were transduced using a firefly-luciferase (luc) and enhanced green fluorescent protein-expressing lentiviral construct as described before.42 The obtained RPMI-8226 luc+ cells were cultured as mentioned above.
In vivo αJAM-A moAb treatment of MM-bearing mice
Eight- to ten-week-old female NSG mice were used in experiments (Charles River, Sulzfeld, Germany). Overall, 2 × 105 RPMI-8226 luc+ cells were suspended in phosphate-buffered saline (PBS) and injected intratibially. One day after the injection, animals were randomized into three groups and imaged by in vivo bioluminescence (BLI) to confirm MM engraftment. Treatment with αJAM-A monoclonal antibody (moAb) started 1 day after MM cell injection. Mice were treated with α-JAM-A moAb 100 μg per 20 g body weight (in 100 μl PBS), mouse monoclonal clone J10.4, purified (Sigma-Aldrich) or an isotype control Ab (mouse IgG Polyclonal Antibody control 12–371, unconjugated, Merck Millipore, Darmstadt, Germany) and 100 μg per 20 g body weight (in 100 μl PBS). The administration schedule was three times per week (nine administrations intraperitonealy) on days 1, 4, 6, 8, 11, 13, 15 and 18; mice were killed for FACS and histology on day 22 after MM injection.
Bioluminescence imaging
BLI was performed on mice with an IVIS Spectrum (Caliper-Xenogen, Alameda, CA, USA) imaging system as previously described.42, 43 Briefly, mice were anesthetized i.p. with a mixture of ketamine (100 mg/kg) and xylazine (10 mg/kg) in PBS. D-Luciferin (150 mg/kg) was co-injected, and BLI measurements were started exactly 10 min later. Details are supplied in the Supplementary Methods.44
Ki-67 and TUNEL staining
The fractions of Ki-67 and TUNEL-positive MM cells were morphologically distinguished from BMSCs by microscopic examination (× 400). Details are described in Supplementary Methods.
Statistical analysis
Continuous data, lacking Gaussian distribution, were summarized as median and interquartile range. Comparison among independent groups was performed by Wilcoxon sum-rank test, whereas relations between variables were explored with Spearman's correlation coefficient.
Qualitative data were summarized as count and percentage, relations and comparisons analyzed with the χ2test. Kaplan–Meier curves were used to show progression-free survival (PFS) and overall survival (OS) related to higher levels of surface JAM-A expression or sJAM-A, and log-rank were determined to evaluate the differences. PFS refers to the time elapsed from the date of enrollment in this study and the date of relapse, disease progression or death, determined from the last follow-up visit (t2). OS refers to the time elapsed from the date of enrollment in this study and date of death from any cause.
Univariate and multivariate Cox models were built to evaluate significant risk factors for OS and PFS in the patient cohort. All analyses were performed with SAS 9.4 for PC Windows. Kaplan–Meier curves were drawn with MedCalc Statistical Software version 17.2 (MedCalc Software bvba, Ostend, Belgium; https://www.medcalc.org; 2017).
For the in vitro experiments Student’s t-test and analysis of variance were used to compare normally distributed data, and Kruskal–Wallis and Mann–Whitney test for non-parametric analyses. Error bars are represented as s.d. Two-way analysis of variance, multiple t-test and Mann–Whitney test were used for in vivo data analysis with GraphPad Prism 6 software (GraphPad Software Inc., La Jolla, CA, USA). A two-sided P-value<0.05 was considered statistically significant.
Results
Decreased PFS in MM patients with elevated JAM expression
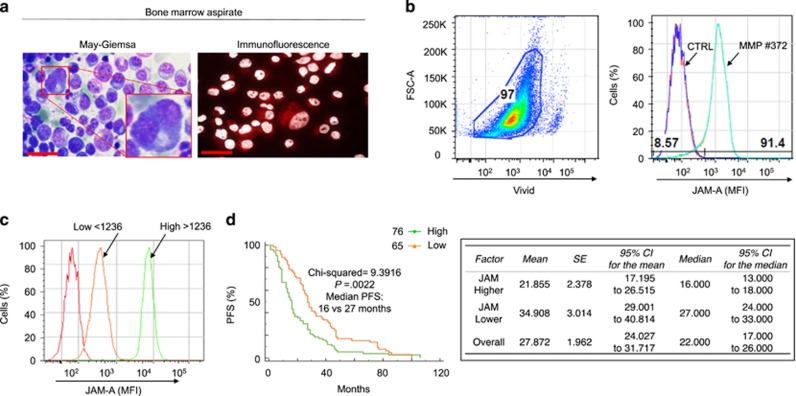
An aberrant expression of JAM family members has been discovered in several tumors. Although the role of JAM-A in carcinogenesis has been subjected to more experimental studies in solid tumors, data about the hemato-oncological clinical significance of the role of JAM-A remain elusive. Therefore, we analyzed the expression and distribution of JAM-A in MM-PCs (Figures 1a and b, representative patient), evaluating the relationship between the expression of membrane JAM-A levels, PFS and OS in 141 subjects out of 147 cases (Figures 1c and d, Supplementary Figure 1, Supplementary Tables 3 and 4): median PFS differed significantly in subjects with higher membrane JAM-A expression levels than the median: in this group PFS was 16 months (95% confidence interval (CI) 13–18 months), whereas PFS in patients with lower membrane JAM-A expression levels reached 27 months (95% CI 24–33). This difference was deemed statistically significant (log-rank=9.3916, P=0.0022). FACS analysis was confirmed at the same time points by immunohistochemistry performed on BM biopsies obtained from patients at the time of recruitment (t0), after therapy (t1) and at disease relapse (t2), as shown in Figure 2. In contrast, PCs obtained from peripheral blood of healthy donors (N=19) or from the MGUS patients BM (MGUS, N=11) did not display detectable JAM-A levels (Supplementary Figure 2A). Multivariate analysis (Supplementary Table 3) confirmed high levels of membrane JAM-A as a statistically significant risk factor for low PFS (hazards ratio (HR)=2.347, 95% CI 1.19–4.608). Similarly, high membrane JAM-A expression levels above the median at remission were correlated with earlier disease progression (HR=1.977, 95% CI 1.141–3.425), as were ISS III (HR=2.319, 95% CI 1.112–4.839).
Figure 1.
JAM-A is overexpressed in MM patient-derived primary plasma cells. (a) MM primary plasma cells (MM-PCs): May–Giemsa and immunofluorescence staining of JAM-A expression from a representative patient. Scale bar, 10 μm. (b) FACS analysis of cell JAM-A surface expression from representative patient-derived, immunoselected MM-PCs, gated on live cells, identified as CD138+ cells. Red and dark blue curves indicate isotype staining and unstained controls. (c) Representative patients with low versus high JAM-A level (FACS). (d) Patients with low expression had a better PFS than patients with high JAM-A expression (log-rank test—left panel). Detailed information for PFS of patients with JAM-A surface expression (right panel). For 5 out of 147 patients JAM-A surface expression was unobtainable and for 1 patient complete information regarding date of disease progression and death was not available.
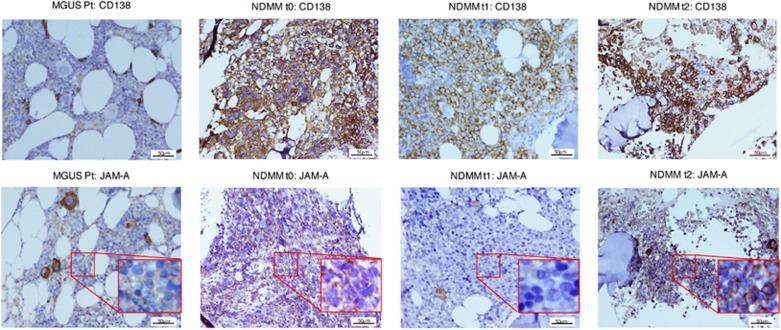
Figure 2.
JAM-A is overexpressed in bone marrow biopsies of MM patients. JAM-A immunohistochemical staining of BM trephines from a single representative MM patient (at t0, t1 and t2) compared with MGUS control. Depending on the disease stage, different JAM-A expression patterns become overt: weak JAM-A staining in MGUS samples, JAM-A expression detectable at the time of recruitment in NDMM, weak after therapy but even stronger at disease relapse. MGUS Pt=MGUS-patient, NDMM=newly diagnosed MM patient. t0=at the time of recruitment; t1=after treatment; t2=at disease relapse. Original magnification × 200, scale bar, 50 μm.
To better address the relation of JAM-A MM-PC surface expression to survival, we compared median OS in the two groups with high and low membrane JAM-A expression levels (Supplementary Figure 1). Subjects with high levels of membrane JAM-A reached a median survival of 53 months (95% CI 48–72) in contrast to the median survival of 60 months (95% CI 48–67) in patients with lower JAM-A levels (log-rank=0.04441, P=0.8331). Nevertheless, the only statistically significant risk factor that affects OS after multivariate analysis seems to be ISS III, that resulted in an HR 2.327 (95% CI 1.112–4.869—Supplementary Table 4).
Subsequently, we measured sJAM-A in the sera obtained from the same subjects. The circulating sJAM-A levels of the MM patients were markedly increased and the average value in MM patients significantly exceeded levels in controls (Supplementary Figure 2B). Nevertheless, multivariate analysis showed no correlation with PFS and OS (Supplementary Tables 3 and 4). The median follow-up was 60 months (95% CI 48–65 months).
Conclusively, MM-PCs JAM-A surface expression was correlated with PFS at both univariate and multivariate analyses and predicted early disease progression.
JAM-A overexpression is correlated with aggressive disease
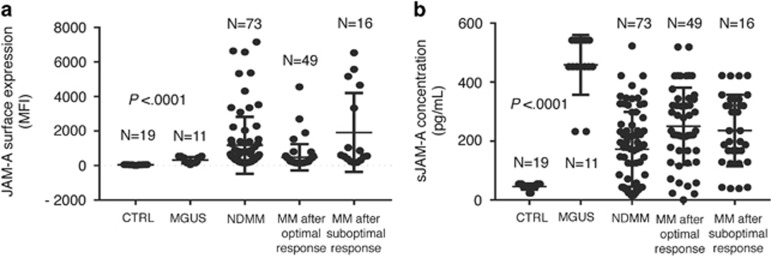
To further explore the relationship between JAM-A expression and MM disease, we investigated the correlation between membrane JAM-A expression and clinicopathological parameters. Therefore, we analyzed the expression and distribution of JAM-A in MM-PCs derived from separate subgroups from our cohort (Figure 3a) and at disease relapse (Supplementary Figure 3A and Supplementary Tables 3–6). JAM-A surface expression in all MM subgroups exceeded expression levels of controls (P<0.0001) and membrane JAM-A was significantly higher at relapse (t2) than after treatment (t1; P<0.0001). Moreover, JAM-A surface level at relapse showed a significant direct correlation with the proportion of BM-PCs' infiltration (rs=0.25; P=0.0051) and with B2-MGB (rs=0.322, P=0.0039; Supplementary Figure 3A, Supplementary Table 5). Remarkably, the membrane JAM-A class (higher than the median, lower than the median) was significantly correlated with ISS (Supplementary Table 6): the percentage of high class of JAM-A at the day of biopsy was 76% in ISS III, 43.4% in ISS II and 45.9% in ISS I (χ2=8.09, P=0.0175), versus 88% in ISS III versus 57.1% in ISS II and 36.7% in ISS I at relapse (χ2= 4.02, P<0.0001). A similar phenomenon was observed for the genetic risk (Supplementary Table 6): 70.8% of genetic high-risk patients had membrane JAM-A levels above the median at the day of biopsy; this percentage decreased in the intermediate risk group to 46.2% and among the standard genetic risk patients to 34% (χ2=14.02, P=0.0009). The same situation was found at relapse, with 89.6% of the subjects at high genetic risk being assigned to the higher membrane JAM-A value class, whereas this class of membrane JAM-A included 34.2% of those in the intermediate and 28.6% in the standard genetic risk groups (χ2=14.45, P<0.0001).
Figure 3.
JAM-A overexpression correlates with aggressive disease. (a) t0: differences in the expression of JAM-A between MM-PCs from NDMM, MM patients after optimal or suboptimal response and PCs obtained from healthy adults and MGUS patients determined by FACS (Kruskal–Wallis test). (b) t0: differences in the serum sJAM-A concentration between NDMM, MM patients after optimal or suboptimal response and serum obtained from healthy adults and MGUS patients. Determined by ELISA (Kruskal–Wallis test). t0=at time of recruitment.
On the basis of univariate analysis (Supplementary Table 5), a high expression of membrane JAM-A at the time of recruitment was significantly inversely correlated with age (rs=−0.29, P=0.0004) and Bence–Jones protein (rs=−0.21, P=0.0298).
All the MM subgroups showed high sJAM-A levels (Figure 3b and Supplementary Tables 5 and 6) and sJAM-A at disease relapse was significantly correlated with the number of bone lesions at the time of recruitment (rs=0.224, P=0.0066) and inversely correlated with paraprotein at disease relapse (rs=−0.318, P=0.0008), and total protein at the time of recruitment (rs=−0.21, P=0.0108). Furthermore, sJAM-A levels at relapse were significantly directly correlated with BM-PCs' infiltration (rs=0.21, P=0.0192, Supplementary Figure 3B) and inversely correlated with BJ (−0.327, P=0.0008) and the number of bone lesions (rs=−0194, P=0.0191). Serum JAM-A at the time of recruitment was directly correlated with disease stage (χ2=9.29, P=0.0096); moreover, sJAM-A after treatment was directly correlated only with ISS: sJAM-A values above the median were present in 48% subjects with ISS III, in 24.5% subjects with ISS II and in 22.9% subjects with ISS I (χ2=6.04, P=0.0488; Supplementary Table 6).
JAM-A inhibition decreases MM cell migration, proliferation and survival in vitro
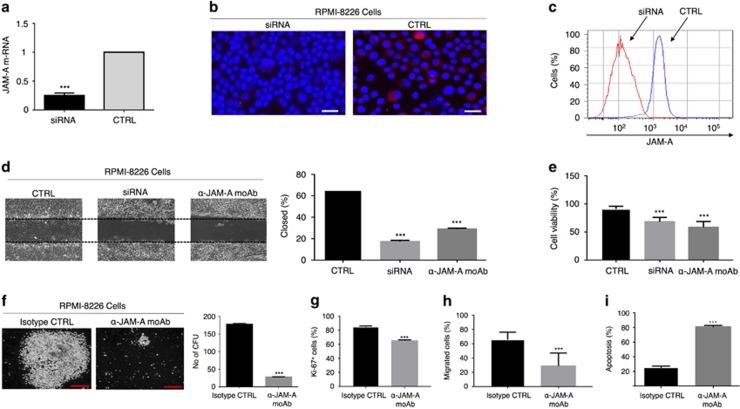
To investigate the clinical findings from primary samples, we investigated JAM-A function in the biology of MM in vitro, particularly regarding MM progression. First, we characterized JAM-A expression in different MM cell lines (RPMI-8226, U266, NCI-H929, OPM-2 and INA-6), at transcriptional level. As controls, HUVEC cells, which express JAM-A upon treatment with TNFα,45 were used. Two of the five MM cell lines homogenously expressed JAM-A (RPMI-8226, U266, P<0.0001), whereas the others expressed little or no JAM-A transcripts (Supplementary Figure 4A). Immunofluorescence microscopy, flow cytometry and western blotting confirmed these results (Supplementary Figures 4B–D). Consequently, we used RPMI-8226 cells in further loss-of-function studies. Using siRNA to knockdown JAM-A expression in this cell line, we addressed whether JAM-A participates in modulating MM cell proliferation and dissemination. JAM-A-specific siRNA effectively suppressed total JAM-A expression, evaluated at both transcriptional and translational levels, as compared with negative control siRNA-transfected cells (Figures 4a–c). Subsequently, we conducted five different functional assays to assess the impact of JAM-A targeting in vitro (Figures 4d–h). Of note, JAM-A knockdown and treatment with an αJAM-A-blocking moAb efficiently reduced the MM cell migration capacity (scratch assay, Figure 4d, CTRL: 61.95±0.95% versus siRNA: 17.78±1.82% and αJAM-A: 29.33±1.74% P<0.0001). JAM-A depletion impaired MM cell viability (Figure 4e, CTRL: 91.85% versus siRNA: 67.87 and αJAM-A: 63.42%. P<0.0001), and colony formation (Figure 4f; negative control versus αJAM-A-blocking moAb, RPMI-8226∶ 271±13 versus 57±5, P<0.0001). Moreover, JAM-A inhibition had an impact on proliferation (Figure 4g, isotype versus αJAM-A moAb=84.67±0.56% versus 65±0.58% 95% CI 21.46–17.88, P<0.0001) and chemotaxis (Figure 4h, isotype versus αJAM-A moAb=67% versus 24.5%, P<0.0001). Finally, αJAM-A moAb treatment determined apoptosis (Figure 4i, isotype versus αJAM-A moAb=28±0.58% versus 78±0.57% 95% CI 48.18−51.82%, P<0.0001).
Figure 4.
JAM-A inhibition impairs key tumorigenic functions of MM cells in vitro. (a) The relative quantity of JAM-A mRNA normalized to GAPDH in the siRNA-transfected group was 75% lower compared with the negative controls (Student’s t-test). (b) Immunofluorescence analysis of the JAM-A signal in RPMI-8226 cells after siRNA treatment. Scale bar, 20 μm. (c) FACS analysis of JAM-A surface expression depletion efficiency. (d) Scratch assay. siRNA- and αJAM-A moAb treatment reduced the capacity of MM cell migration (one-way analysis of variance (ANOVA)). (e) Cell survival function assessed by trypan blue exclusion assay. JAM-A impairs cell viability (Kruskal–Wallis test). (f) Colony-forming unit assay. αJAM-A moAb treatment reduced the ability of cell colony formation when compared with treatment with an isotype control. Scale bar, 100 μm (Student’s t-test). Data are presented as mean±s.d. of triplicate cultures (g) Ki-67 proliferation assay. αJAM-A moAb treatment reduces cell proliferation in treated cells compared with treatment with an isotype control (one-way ANOVA). (h) Chemotaxis assay. αJAM-A moAb treatment reduces the chemotaxis ability of the cells compared with treatment with an isotype control (Kruskal–Wallis test). (i) Effect of JAM-A on cell death evaluated with Annexin (Student’s t-test). ***P<0.0001.
αJAM-A moAb treatment inhibits MM progression in vivo
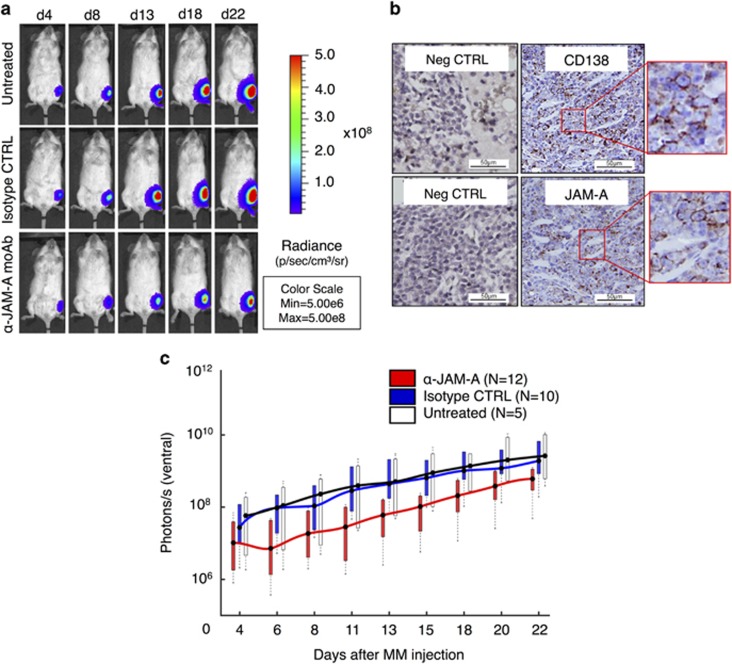
On the basis of our in vitro observations, we examined whether blocking JAM-A would impair MM disease progression in vivo. Therefore, we treated MM-bearing mice with αJAM-A moAb and monitored disease progression with BLI. To address treatment response on established MM, we treated mice over the course of 20 days with either a blocking anti-JAM-A moAb (‘αJAM-A moAb’ N=12), an IgG1 isotype control moAb (‘Isotype CTRL’ N=10) or left them untreated (N=5; Figure 5a). Histologic analysis of tumor-bearing mice confirmed the results of the BLI imaging (Figure 5b). The RPMI-8226 luc+ cells infiltrated the BM in many small clusters, comprising 70% of cellular elements in total. BM analysis revealed CD138+JAM-A+ double-positive cells (Figure 5b). BLI signals increased more strongly in isotype CTRL and untreated animals than in αJAM-A moAb-treated mice (Figure 5c). Treatment showed an overall significant difference in MM progression when comparing the three experimental groups (P=0.0189). The first significant differences were reached on day 8 after MM cell injection, when comparing JAM-A treated with isotype-treated mice (P=0.047) as well as with untreated mice (P=0.028). MM progression in isotype-treated mice did not differ statistically at any time point from untreated mice. Ex vivo BLI imaging of the tibia from tumor-bearing mice revealed a strong infiltration of MM cells into the BM (data not shown). Furthermore, sJAM-A serum levels from MM-bearing mice significantly exceeded those of healthy mice on day 22 (275±27.62 pg/ml, N=8 versus 14.81±3.53 pg/ml, N=3; P<0.0003, Supplementary Figure 5).
Figure 5.
In vivo MM progression is inhibited by αJAM-A moAb treatment. (a) Blocking JAM-A impairs proliferation of luc+ RMPI-8226 cells in vivo. In vivo BLI imaging from RPMI-8226 tumor-bearing mice. Images show ventral views of one representative mouse from each group. (b) Histologic analysis reveals CD138/JAM-A double-positive cells (right panels and magnified boxes) within the BM localized as small clusters and comprising ~70% of cellular elements in total, as analyzed by two independent pathologists. Control staining with monoclonal isotype antibody (Neg. CTRL, left panels). Scale bar, 50 μm. (c) Absolute light-emission (photons/s/animal) during the treatment period. BLI signals markedly increased in control animals (isotype and untreated). αJAM-A moAb treatment impaired disease progression, with the first significant differences appearing on day 8 after MM cell injection (one-way ANOVA; P=0.014) and dorsal (iso-CTRL versus αJAM-A P<0.0001) images.
Discussion
Upregulation of JAM-A has previously been linked to cancer progression.29, 30, 31, 32, 33 Collectively, our results show, for the first time that human JAM-A controls tumor proliferation in vitro and in vivo. Therefore, we postulate that JAM-A may be a potential novel target against MM.
The moAb J10.4, by specifically targeting JAM-A, decreased tumor activities in vitro and disease progression in vivo and exerted significant antitumor effects in xenograft models of human tumors. The present study offers new insights into a potential double role for adhesion molecules like JAM-A, as clinically significant biomarkers and as novel therapeutic targets in MM.
Given the findings described in solid tumors, JAM-A expression seems to promote proliferation and inhibit apoptosis.20 So far, little is known about the role JAM-A may have in hematological malignancies, whereas a role for JAM-A as a proliferation enhancer has been highlighted by in vivo studies in which JAM-A inhibition led to tumor shrinkage in breast, head and neck, and lung cancer models, due to decreased proliferation.29, 30, 31, 32 The dependence of these events on JAM-A dimerization20, 27 might suggest an alternative regulation of JAM-A signaling between physiological versus pathophysiological settings. It is intriguing to speculate that in the physiological setting of an intact epithelial barrier, JAM-A homodimerization between adjacent cells could inhibit proliferation to maintain barrier homeostasis. In contrast, in pathophysiological settings, for example, JAM-A upregulation in cancer,27 JAM-A overexpression distributed across the surface of less-polarized cells might engage in adhesion-independent signaling to promote cell proliferation. Overall, this suggests a complex spatial and temporal regulation by JAM-A of the cellular phenotype. This offers great promise for selective pharmacological targeting of JAM-A. Interestingly, a study published while our data were being concluded39 found JAM-A to be a predictive biomarker for sensitivity to the Reolysin-induced cell death in advanced MM. Our study substantially extends these findings by providing a deeper insight into the translational relevance of therapeutic JAM-A targeting. Furthermore, we also show a prognostic significance of JAM-A expression for MM patient. In the current study, MM patients with poor prognosis displayed significantly higher JAM-A cell surface and serum levels. Our findings should encourage a prospective survival study with higher number of patients to clarify the correlation of MM-PCs' JAM-A surface expression with OS and JAM-A serum levels' relationship with OS and PFS. Unlike the membrane expression of the molecule, a relatively specific indicator of the JAM-A tumor load, the JAM-A serum concentration reflects the production and secretion, which depend on several cell-intrinsic, local and systemic factors such as inflammation and angiogenesis.31 Conclusively, we propose JAM-A as a promising diagnostic and therapeutic target in MM.
Acknowledgments
This work was supported by the University of Bari ‘Aldo Moro’—Internal Medicine Residency Program; DFG FOR 1586 SKELMET; DFG KFO 216 and the Interdisziplinäres Zentrum für Klinische Forschung (IZKF-B233) of the University of Würzburg. We thank Sina Thusek, all members of the Beilhack lab for technical support and fruitful discussions and Mary Victoria Pragnell, BA from the School of Medicine and Surgery, University of Bari for linguistic editing.
Footnotes
Supplementary Information accompanies this paper on the Leukemia website (http://www.nature.com/leu)
The authors declare no conflict of interest.
Supplementary Material
References
- Cavo M, Rajkumar SV, Palumbo A, Moreau P, Orlowski R, Bladé J et al. International Myeloma Working Group consensus approach to the treatment of multiple myeloma patients who are candidates for autologous stem cell transplantation. Blood 2011; 117: 6063–6073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikhael JR, Dingli D, Roy V, Reeder CB, Buadi FK, Hayman SR et al. Management of newly diagnosed symptomatic multiple myeloma: updated Mayo Stratification of Myeloma and Risk-Adapted Therapy (mSMART) consensus guidelines 2013. Mayo Clin Proc 2013; 88: 360–376. [DOI] [PubMed] [Google Scholar]
- Fairfield H, Falank C, Avery L, Reagan MR. Multiple myeloma in the marrow: pathogenesis and treatments. Ann N Y Acad Sci 2016; 1364: 32–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damiano JS, Cress AE, Hazlehurst LA, Shtil AA, Dalton WS. Cell adhesion mediated drug resistance (CAM-DR): role of integrins and resistance to apoptosis in human myeloma cell lines. Blood 1999; 93: 1658–1667. [PMC free article] [PubMed] [Google Scholar]
- Mutlu P, Ural AU, Gunduz U. Differential gene expression analysis related to extracellular matrix components in drug-resistant RPMI-8226 cell line. Biomed Pharmacother Biomed Pharmacother 2012; 66: 228–231. [DOI] [PubMed] [Google Scholar]
- Furukawa Y, Kikuchi J. Epigenetic mechanisms of cell adhesion-mediated drug resistance in multiple myeloma. Int J Hematol 2016; 104: 281–292. [DOI] [PubMed] [Google Scholar]
- Di Marzo L, Desantis V, Solimando AG, Ruggieri S, Annese T, Nico B et al. Microenvironment drug resistance in multiple myeloma: emerging new players. Oncotarget 2016; 26: 60698–60711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chauhan D, Uchiyama H, Akbarali Y, Urashima M, Yamamoto K, Libermann TA et al. Multiple myeloma cell adhesion-induced interleukin-6 expression in bone marrow stromal cells involves activation of NF-kappa B. Blood 1996; 87: 1104–1112. [PubMed] [Google Scholar]
- Hideshima T, Bergsagel PL, Kuehl WM, Anderson KC. Advances in biology of multiple myeloma: clinical applications. Blood 2004; 104: 607–618. [DOI] [PubMed] [Google Scholar]
- Noborio-Hatano K, Kikuchi J, Takatoku M, Shimizu R, Wada T, Ueda M et al. Bortezomib overcomes cell-adhesion-mediated drug resistance through downregulation of VLA-4 expression in multiple myeloma. Oncogene 2009; 28: 231–242. [DOI] [PubMed] [Google Scholar]
- Pulido R, Elices MJ, Campanero MR, Osborn L, Schiffer S, Garcia-Pardo A et al. Functional evidence for three distinct and independently inhibitable adhesion activities mediated by the human integrin VLA-4. Correlation with distinct alpha 4 epitopes. J Biol Chem 1991; 266: 10241–10245. [PubMed] [Google Scholar]
- Gutierrez NC, Garcia JL, Hernandez JM, Lumbreras E, Castellanos M, Rasillo A et al. Prognostic and biologic significance of chromosomal imbalances assessed by comparative genomic hybridization in multiple myeloma. Blood 2004; 104: 2661–2666. [DOI] [PubMed] [Google Scholar]
- Hazlehurst LA, Damiano JS, Buyuksal I, Pledger WJ, Dalton WS. Adhesion to fibronectin via beta1 integrins regulates p27kip1 levels and contributes to cell adhesion mediated drug resistance (CAM-DR). Oncogene 2000; 19: 4319–4327. [DOI] [PubMed] [Google Scholar]
- Neri P, Ren L, Azab AK, Brentnall M, Gratton K, Klimowicz AC et al. Integrin beta7-mediated regulation of multiple myeloma cell adhesion, migration, and invasion. Blood 2011; 117: 6202–6213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Vuori K, Reed JC, Ruoslahti E. The alpha 5 beta 1 integrin supports survival of cells on fibronectin and up-regulates Bcl-2 expression. Proc Natl Acad Sci USA 1995; 92: 6161–6165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paiva B, Corchete LA, Vidriales MB, Puig N, Maiso P, Rodriguez I et al. Phenotypic and genomic analysis of multiple myeloma minimal residual disease tumor cells: a new model to understand chemoresistance. Blood 2016; 127: 1896–1906. [DOI] [PubMed] [Google Scholar]
- Nefedova Y, Cheng P, Alsina M, Dalton WS, Gabrilovich DI. Involvement of Notch-1 signaling in bone marrow stroma-mediated de novo drug resistance of myeloma and other malignant lymphoid cell lines. Blood 2004; 103: 3503–3510. [DOI] [PubMed] [Google Scholar]
- Tamura H, Ishibashi M, Yamashita T, Tanosaki S, Okuyama N, Kondo A et al. Marrow stromal cells induce B7-H1 expression on myeloma cells, generating aggressive characteristics in multiple myeloma. Leukemia 2013; 27: 464–472. [DOI] [PubMed] [Google Scholar]
- Podar K, Zimmerhackl A, Fulciniti M, Tonon G, Hainz U, Tai YT et al. The selective adhesion molecule inhibitor Natalizumab decreases multiple myeloma cell growth in the bone marrow microenvironment: therapeutic implications. Br J Haematol 2011; 155: 438–448. [DOI] [PubMed] [Google Scholar]
- Dotterweich J, Tower RJ, Brandl A, Muller M, Hofbauer LC, Beilhack A et al. The KISS1 receptor as an in vivo microenvironment imaging biomarker of multiple myeloma bone disease. PLoS One 2016; 11: e0155087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Julia D, Katrin S, Alexander K, Beate G, Doris S, Sabine Z et al. Contact of myeloma cells induces a characteristic transcriptome signature in skeletal precursor cells—implications for myeloma bone disease. Bone 2016; 93: 155–166. [DOI] [PubMed] [Google Scholar]
- Philipp-Abbrederis K, Herrmann K, Knop S, Schottelius M, Eiber M, Luckerath K et al. In vivo molecular imaging of chemokine receptor CXCR4 expression in patients with advanced multiple myeloma. EMBO Mol Med 2015; 7: 477–487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang C-H, Hale SJ, Cox CV, Blair A, Kronsteiner B, Grabowska R et al. Junctional adhesion molecule-A is highly expressed on human hematopoietic repopulating cells and associates with the key hematopoietic chemokine receptor CXCR4. Stem Cells 2016; 34: 1664–1678. [DOI] [PubMed] [Google Scholar]
- Kobayashi I, Kobayashi-Sun J, Kim AD, Pouget C, Fujita N, Suda T et al. Jam1a-Jam2a interactions regulate haematopoietic stem cell fate through Notch signalling. Nature 2014; 512: 319–323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin-Padura I, Lostaglio S, Schneemann M, Williams L, Romano M, Fruscella P et al. Junctional adhesion molecule, a novel member of the immunoglobulin superfamily that distributes at intercellular junctions and modulates monocyte transmigration. J Cell Biol 1998; 142: 117–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leech AO, Cruz RG, Hill AD, Hopkins AM. Paradigms lost-an emerging role for over-expression of tight junction adhesion proteins in cancer pathogenesis. Ann Transl Med 2015; 3: 184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugano Y, Takeuchi M, Hirata A, Matsushita H, Kitamura T, Tanaka M et al. Junctional adhesion molecule-A, JAM-A, is a novel cell-surface marker for long-term repopulating hematopoietic stem cells. Blood 2008; 111: 1167–1172. [DOI] [PubMed] [Google Scholar]
- Weber C, Fraemohs L, Dejana E. The role of junctional adhesion molecules in vascular inflammation. Nat Rev Immunol 2007; 7: 467–477. [DOI] [PubMed] [Google Scholar]
- Scott DW, Tolbert CE, Burridge K. Tension on JAM-A activates RhoA via GEF-H1 and p115 RhoGEF. Mol Biol Cell 2016; 27: 1420–1430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Severson EA, Lee WY, Capaldo CT, Nusrat A, Parkos CA. Junctional adhesion molecule A interacts with Afadin and PDZ-GEF2 to activate Rap1A, regulate beta1 integrin levels, and enhance cell migration. Mol Biol Cell 2009; 20: 1916–1925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koenen RR, Pruessmeyer J, Soehnlein O, Fraemohs L, Zernecke A, Schwarz N et al. Regulated release and functional modulation of junctional adhesion molecule A by disintegrin metalloproteinases. Blood 2009; 113: 4799–4809. [DOI] [PubMed] [Google Scholar]
- Ong KL, Leung RY, Babinska A, Salifu MO, Ehrlich YH, Kornecki E et al. Elevated plasma level of soluble F11 receptor/junctional adhesion molecule-A (F11R/JAM-A) in hypertension. Am J Hypertens 2009; 22: 500–505. [DOI] [PubMed] [Google Scholar]
- Zhao C, Lu F, Chen H, Zhao X, Sun J, Chen H. Dysregulation of JAM-A plays an important role in human tumor progression. Int J Clin Exp Pathol 2014; 7: 7242–7248. [PMC free article] [PubMed] [Google Scholar]
- Zhang M, Luo W, Huang B, Liu Z, Sun L, Zhang Q et al. Overexpression of JAM-A in non-small cell lung cancer correlates with tumor progression. PLoS One 2013; 8: e79173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian Y, Tian Y, Zhang W, Wei F, Yang J, Luo X et al. Junctional adhesion molecule-A, an epithelial-mesenchymal transition inducer, correlates with metastasis and poor prognosis in human nasopharyngeal cancer. Carcinogenesis 2015; 36: 41–48. [DOI] [PubMed] [Google Scholar]
- Lathia JD, Li M, Sinyuk M, Alvarado AG, Flavahan WA, Stoltz K et al. High-throughput flow cytometry screening reveals a role for junctional adhesion molecule a as a cancer stem cell maintenance factor. Cell Rep 2014; 6: 117–129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kakuki T, Kurose M, Takano KI, Kondoh A, Obata K, Nomura K et al. Dysregulation of junctional adhesion molecule-A via p63/GATA-3 in head and neck squamous cell carcinoma. Oncotarget 2016; 28: 33887–33900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goetsch L, Haeuw JF, Beau-Larvor C, Gonzalez A, Zanna L, Malissard M et al. A novel role for junctional adhesion molecule-A in tumor proliferation: modulation by an anti-JAM-A monoclonal antibody. Int J Cancer 2013; 132: 1463–1474. [DOI] [PubMed] [Google Scholar]
- Kelly KR, Espitia CM, Zhao W, Wendlandt E, Tricot G, Zhan F et al. Junctional adhesion molecule-A is overexpressed in advanced multiple myeloma and determines response to oncolytic reovirus. Oncotarget 2015; 6: 41275–41289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burger R, Guenther A, Bakker F, Schmalzing M, Bernand S, Baum W et al. Gp130 and ras mediated signaling in human plasma cell line INA-6: a cytokine-regulated tumor model for plasmacytoma. Hematol J 2001; 2: 42–53. [DOI] [PubMed] [Google Scholar]
- Ikegame A, Ozaki S, Tsuji D, Harada T, Fujii S, Nakamura S et al. Small molecule antibody targeting HLA class I inhibits myeloma cancer stem cells by repressing pluripotency-associated transcription factors. Leukemia 2012; 26: 2124–2134. [DOI] [PubMed] [Google Scholar]
- Thalheimer A, Korb D, Bonicke L, Wiegering A, Muhling B, Schneider M et al. Noninvasive visualization of tumor growth in a human colorectal liver metastases xenograft model using bioluminescence in vivo imaging. J Surg Res 2013; 185: 143–151. [DOI] [PubMed] [Google Scholar]
- Riedel SS, Mottok A, Brede C, Bauerlein CA, Jordan Garrote AL, Ritz M et al. Non-invasive imaging provides spatiotemporal information on disease progression and response to therapy in a murine model of multiple myeloma. PLoS One 2012; 7: e52398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brede C, Friedrich M, Jordan-Garrote AL, Riedel SS, Bauerlein CA, Heinze KG et al. Mapping immune processes in intact tissues at cellular resolution. J Clin Invest 2012; 122: 4439–4446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozaki H, Ishii K, Horiuchi H, Arai H, Kawamoto T, Okawa K et al. Cutting edge: combined treatment of TNF-alpha and IFN-gamma causes redistribution of junctional adhesion molecule in human endothelial cells. J Immunol 1999; 163: 553–557. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.