Abstract
Background
Rhamnolipids are the most extensively studied biosurfactants and has been successfully used in various areas from bioremediation to industrial fields. Rhamnolipids structural composition decide their physicochemical properties. Different physicochemical properties influence their application potential. Rhamnolipids can be produced at both aerobic conditions and anaerobic conditions by Pseudomonas aeruginosa. This study aims to evaluate the oxygen effects on the rhamnolipids yield, structural composition, physicochemical properties and the rhl-genes expression in P. aeruginosa SG. Results will guide researchers to regulate microbial cells to synthesize rhamnolipids with different activity according to diverse application requirements.
Results
Quantitative real-time PCR analysis revealed that rhlAB genes were down-regulated under anaerobic conditions. Therefore, strain P. aeruginosa SG anaerobically produced less rhamnolipids (0.68 g/L) than that (11.65 g/L) under aerobic conditions when grown in media containing glycerol and nitrate. HPLC–MS analysis showed that aerobically produced rhamnolipids mainly contained Rha-C8-C10, Rha–Rha-C10-C12:1 and Rha–Rha-C8-C10; anaerobically produced rhamnolipids mainly contained Rha-C10-C12 and Rha-C10-C10. Anaerobically produced rhamnolipids contained more mono-rhamnolipids (94.7%) than that (54.8%) in aerobically produced rhamnolipids. rhlC gene was also down-regulated under anaerobic conditions, catalyzing less mono-rhamnolipids to form di-rhamnolipids. Aerobically produced rhamnolipids decreased air–water surface tension (ST) from 72.2 to 27.9 mN/m with critical micelle concentration (CMC) of 60 mg/L; anaerobically produced rhamnolipids reduced ST to 33.1 mN/m with CMC of 80 mg/L. Anaerobically produced rhamnolipids emulsified crude oil with EI24 = 80.3%, and aerobically produced rhamnolipids emulsified crude oil with EI24 = 62.3%. Both two rhamnolipids products retained surface activity (ST < 35.0 mN/m) and emulsifying activity (EI24 > 60.0%) under temperatures (4–121 °C), pH values (4–10) and NaCl concentrations less than 90 g/L.
Conclusions
Oxygen affected the rhl-genes expression in P. aeruginosa, thus altering the rhamnolipids yield, structural composition and physicochemical properties. Rhamnolipids produced at aerobic or anaerobic conditions was structurally distinct. Two rhamnolipids products had different application potential in diverse biotechnologies. Although both rhamnolipids products were thermo-stable and halo-tolerant, aerobically produced rhamnolipids possessed better surface activity, implying its well wetting activity and desorption property; anaerobically produced rhamnolipids exhibited better emulsifying activity, indicating its applicability for enhanced oil recovery and bioremediation of petroleum pollution.
Keywords: Mono-rhamnolipids, Di-rhamnolipids, HPLC–MS, rhl-genes, Emulsifying activity, Pseudomonas aeruginosa
Background
Rhamnolipids are one of the most popular biosurfactants in current research [1, 2]. Due to their high surface activity and emulsifying activity, solubilization activity, low toxicity, and biodegradability [3, 4], rhamnolipids have been broadly applied in many different fields, such as microbial enhanced oil recovery (MEOR) [5–7], bioremediation of hydrocarbon pollutants and heavy metals [8–10].
Rhamnolipids are mainly produced by Pseudomonas aeruginosa [11, 12]. P. aeruginosa is one of facultative bacteria, which can grow and metabolize in both aerobic and anaerobic environments [13, 14]. Almost all studies about rhamnolipids production by P. aeruginosa were focused on aerobic conditions. However, some P. aeruginosa strains can also produce rhamnolipids under anaerobic conditions [15, 16]. What is the difference between aerobic production of rhamnolipids and anaerobic production of rhamnolipids? Information of oxygen effects (aerobic cultivation and anaerobic cultivation) on the rhamnolipids production is still scarce.
Rhamnolipids contain one or two rhamnoses attached to one or two β-hydroxyl fatty acids [11, 17]. Besides, the β-hydroxyl fatty acids chains vary significantly with lengths of 8, 10, 12, and 14 carbons [17]. In the rhamnolipids biosynthesis process, these groups may be randomly grouped to form different rhamnolipids homologues. Therefore, the structural composition of rhamnolipids are highly diverse [11].
The same P. aeruginosa strain can produce diverse rhamnolipids mixtures under different culture conditions. Déziel et al. reported strain P. aeruginosa 57RP produced different rhamnolipids mixtures grown on mannitol or naphthalene [18]. Nicolò et al. analyzed the different ratio of mono-rhamnolipids to di-rhamnolipids produced by P. aeruginosa L05 using four different carbon source [19]. Aerobic cultivation and anaerobic cultivation would also influence P. aeruginosa to produce rhamnolipids with different structural composition. Rhamnolipids structural composition decide their physicochemical properties. Changes in rhamnolipids composition can affect their properties, such as the CMC and emulsifying activity [20]. The different physicochemical properties of rhamnolipids will influence its application potential in different field. It is significant to understand the relationship between the rhamnolipids composition and its physicochemical activities, such as surface activity, emulsifying activity. The aim of this work was to investigate the effect of oxygen conditions (aerobic cultivation and anaerobic cultivation) on the rhamnolipids structural composition and physicochemical properties. Results will guide us to regulate microorganisms to synthesize highly active rhamnolipids according to different biotechnological application requirements.
In P. aeruginosa, the rhlA and rhlB genes arranged in an operon code the rhamnosyltransferase-1 (RhlAB) [21]. RhlA is involved in the synthesis of β-hydroxyl fatty acids. RhlB catalyzes dTDP-l-rhamnose and β-hydroxyl fatty acids to synthesis mono-rhamnolipids. The rhlC gene codes the rhamnosyltransferase-2 (RhlC) that catalyzes the second dTDP-l-rhamnose molecule adding to mono-rhamnolipids to form di-rhamnolipids [17]. In order to analyze the mechanism of oxygen effects on rhamnolipids production, expression of rhlAB genes and rhlC gene in P. aeruginosa under aerobic and anaerobic conditions was studied by quantitative real-time PCR (qRT-PCR).
In the present study, strain P. aeruginosa SG was used to produce rhamnolipids under aerobic or anaerobic conditions. This aims to evaluate the possible influence of oxygen on the rhamnolipids production in strain P. aeruginosa SG. Rhamnolipids production capacity of strain SG under aerobic conditions or anaerobic conditions was comparatively studied. The structure difference of two rhamnolipids products was analyzed by Fourier Transform infrared spectroscopy (FTIR) and high performance liquid chromatography–mass spectrometry (HPLC–MS). The different ratio of mono-rhamnolipids to di-rhamnolipids was also analyzed. The physicochemical properties of two rhamnolipids products were also comparatively evaluated, including surface activity, critical micelle concentration (CMC), emulsifying activity and stability. Application potential of two rhamnolipids products was also discussed.
Methods
Bacterial strain, medium and culture conditions
The bacterial strain P. aeruginosa SG (GenBank accession number KJ995745) isolated from oil reservoir environments was used for production of rhamnolipids under aerobic or anaerobic conditions, respectively [16]. The glycerol–nitrate (GN) medium with some improvements was used for culture of strain SG [22]. The GN medium contains 70.3 g/L of glycerol, 5.2 g/L of NaNO3, 6.9 g/L of K2HPO4·3H2O, 5.5 g/L of KH2PO4, 0.40 g/L of MgSO4∙7H2O, 1.0 g/L of NaCl, 1.0 g/L of KCl, 0.5 g/L of yeast extract and 0.13 g/L of CaCl2. The medium pH was adjusted to 6.8. All the chemicals and solvents (Sinopharm Chemical Reagent, Beijing, China) were analytical grade. The aerobic medium was dispensed into Erlenmeyer flasks and sterilized (121 °C, 20 min). The 500-mL Erlenmeyer flasks containing 250 mL GN medium were used for aerobic cultivation. Aerobic cultivation experiments were performed at 37 °C and 180 rpm for 6 days with three replicate. The anaerobic medium was prepared as follows. The medium was firstly boiled under a stream of oxygen-free N2 gas for 15 min. Then the medium was dispensed into serum bottles under a stream of oxygen-free N2 and sterilized (121 °C, 20 min). The 250-mL serum bottles containing 200 mL anaerobic GN medium were used for anaerobic cultivation. Anaerobic cultivation experiments were performed at 37 °C and 80 rpm for 10 days with three replicate.
Comparative analysis of rhlAB and rhlC genes expression
Expression of the rhlAB genes and rhlC gene was investigated by quantification of mRNA transcripts using qRT-PCR. For total RNA extraction, 1 mL culture was sampled at the stationary growth phase. Because rhamnolipids yield reached maximum at the stationary growth phase. Strain SG was cultivated for 100 h at 37 °C, 180 rpm under aerobic conditions, and for 150 h at 37 °C, 80 rpm under anaerobic conditions, respectively. Cultivation experiments were performed with three replicate. Total RNA of samples was extracted using TaKaRa MiniBEST Universal RNA Extraction Kit (Takara, Japan) according to the manufacturer’s instructions. Random-primed reverse transcription was conducted using PrimeScript™ Double Strand cDNA Synthesis Kit (Takara, Japan) according to the manufacturer’s instructions.
The relative expression values of genes rhlAB and rhlC were obtained by the comparative CT (2−ΔΔCT) method [23–25]. 16S rRNA gene was used as endogenous control. The rhlAB genes, rhlC gene and 16S rRNA gene were amplified and quantified from cDNA using using a LightCycler instrument (Roche) and the SYBR Green I fluorophore protocol. The real-time quantitative PCR primers used for rhlAB genes, rhlC gene and 16S rRNA gene were as follows: rhlAB-F (TCAACGAGACCGTCGGCAAATACCT) and rhlAB-R (AATCCCGTACTTCTCGTGAGCGATG), rhlC-F (ATCCATCTCGACGGACTGAC) and rhlC-R (GTCCAGGTCGTCGATGAAC), 16S-341F (CCTACGGGAGGCAGCAG) and 16S-518R (ATTACCGCGGCTGCTGG). The 20 μL reaction volumes contained 10 μL of SYBR Premix Ex Taq™ II (Tli RNaseH Plus) (Takara, Japan), 0.8 μL of each primer (10 μM), 2 μL of template cDNA and 6.4 μL of PCR-grade water. Adding 2 μL of PCR-grade water instead of DNA template was used as a negative control. All amplifications were performed in triplicate. Thermal cycling consisted of one cycle of 95 °C for 3 min, followed by 40 cycles of 95 °C for 15 s, 55 °C for 30 s, and 72 °C for 45 s, followed by melt curve analysis to confirm the specificity of the amplicons. Fluorescence was measured during the last step of each PCR cycle. The relative expression values of rhlAB genes and rhlC gene were calculated using the comparative CT (2−ΔΔCT) method [23–25]. Relative expression values above and below 1 means a higher and a lower expression level under anaerobic conditions than that under aerobic conditions, respectively.
Rhamnolipids extraction and quantification
The rhamnolipids products were extracted by the method referred to previous studies with some modifications [26, 27]. The bacterial cells were removed by centrifugation (10,000g, 10 min) to obtain supernatant. After heating at 80 °C for 30 min to denature the extracellular proteins, the pH value of supernatant was adjusted to 2.0 using 2 mol/L of HCl-water solution. Then the supernatant was kept at 4 °C for 12 h to precipitate rhamnolipids. Chloroform and methanol organic solvent (v/v, 2:1) was used for rhamnolipids extraction. The lower organic phase was collected and evaporated to dryness using a rotary evaporator (50 rpm, 45 °C). The thin yellowish rhamnolipids product was used for further analysis.
Oil spreading method was used for rhamnolipids quantification [28]. Briefly, 30 mL of distilled water was added into a 90 mm petri dish. Then 20 μL of crude oil was added to the surface of water to form a thin oil membrane. Then 10 μL of sample was gently dropped onto the center of the oil membrane. A clearly oil spreading circle was formed. The diameter of oil spreading circle was measured and recorded. All experiments were conducted three times. According to the diameter of formed oil spreading circle, the amount of rhamnolipids in culture was calculated from standard curves prepared with extracted rhamnolipids (100–800 mg/L). Therefore, samples which contain rhamnolipids high than 800 mg/L need to be diluted. The aerobically produced rhamnolipids extract and the anaerobically produced rhamnolipids extract were, respectively, used to prepare standard curves for quantify rhamnolipids in aerobic culture and anaerobic culture.
Fourier Transform infrared spectroscopy analysis
The FTIR spectra of the aerobically produced rhamnolipids and the anaerobically produced rhamnolipids of strain SG were recorded by a NICOLET 380 FTIR spectrometer. Rhamnolipids product (10 mg) was mixed with 90 mg of KBr (spectral purity). The mixture was pressed with pressure of 25 Mpa for 25 s. The translucent pellet was used for FT-IR analysis with the resolution of 0.5 cm−1 and the wave number range from 400 to 4000 cm−1.
HPLC–MS analysis
The aerobic produced rhamnolipids and the anaerobic produced rhamnolipids were dissolved into 10% acetonitrile (in water) solution containing 2 mM ammonium acetate, respectively. Two rhamnolipids solutions with concentrations of 500 mg/L were prepared for congener compositions analysis by high-performance liquid chromatograph-mass spectrometer (Waters, USA). The HPLC–MS analysis was performed according to Déziel’s method with minor modification [18]. A C18 reverse phase column (150 mm × 2 mm, particle size 5 μm) was used. An acetonitrile–water gradient from 10 to 60% was used as mobile phase. The HPLC flow rate was 0.6 mL/min. 10% of the flow was introduced into the mass spectrometer. Capillary voltage was set at 3.8 kV, cone voltage at 35 V and the source temperature was kept at 100 °C. The mass spectrometer was operated in the negative ion mode scanning 50–800 m/z range. Relative percentage content of each rhamnolipids homologue was calculated by area normalization method.
Determination of critical micelle concentration
Rhamnolipids solutions (concentrations of 0–160 mg/L) were prepared to determine critical micelle concentration (CMC) values of aerobically produced rhamnolipids and the anaerobically produced rhamnolipids. The surface tension of each solution was measured. The relationship graphs between the surface tension and concentration of rhamnolipids were prepared. In the graph, the relevant rhamnolipids concentration at the turning point of surface tension is the CMC of rhamnolipids product, i.e., the surface tension reaches the lowest at the CMC value of rhamnolipids product.
Analysis of surface activity and emulsifying activity
Two rhamnolipids-water solutions (concentrations of 1000 mg/L) were prepared using the aerobically produced rhamnolipids and the anaerobically produced rhamnolipids, respectively. Surface tension of rhamnolipids solutions was measured by a BZY-1 automatic surface tension meter (Shanghai equitable Instruments Factory, china). Emulsification index (EI24) was measured to evaluate emulsifying activity. Briefly, 4 mL of hydrophobic organics was mixed with 4 mL of rhamnolipids solutions in test tubes. The tubes were stirred at a vortex mixer for 2 min and placed at 37 °C for 24 h [16]. The EI24 (%) is the height of the emulsion layer (mm) divided by the total height of the mixture (mm) and multiplied by 100 [16]. The tested hydrophobic organics included kerosene, liquid paraffin, petroleum ether, isooctane, cyclohexane, n-hexane, benzene, olive oil and crude oil (Xinjiang Oilfield, China).
Stability evaluation of two rhamnolipids product
Stability of the aerobically produced rhamnolipids and the anaerobically produced rhamnolipids were evaluated using 1000 mg/L of rhamnolipids solutions. 1 mol/L HCl and 1 mol/L NaOH were used to adjust pH values of samples. A series of 20 mL of two kinds of rhamnolipids solutions were treated under different temperatures (4, 25, 35, 45, 60, 80, 100 and 121 °C), different pH values (2, 4, 6, 8, 10 and 12) and different salinities (NaCl concentrations of 0, 3, 6, 9, 12, 15, 18, 21 and 25%). Surface tension and EI24 index (using crude oil as emulsion phase) were measured at 35 °C to evaluate the stability of the aerobically produced rhamnolipids and the anaerobically produced rhamnolipids.
Results and discussion
Rhamnolipids yield and rhlAB genes expression under two conditions
The diameters of the formed oil spreading circle are positively associated with the rhamnolipid concentrations (100–800 mg/L). For the aerobically produced rhamnolipid, the linear correlation model between the diameters of oil spreading circle (y) and rhamnolipid concentrations (x) was y = 0.0591x + 6.24, R2 = 0.9931. Another positive linear correlation model for the anaerobically produced rhamnolipid was y = 0.0616x + 5.9524, R2 = 0.9908. According to the diameter of formed oil spreading circle, the amount of rhamnolipids in culture was calculated using two positive linear correlation models. Strain SG obtained highest rhamnolipids yield under aerobic conditions at the 5th day. The rhamnolipids concentration in SG aerobic culture was 11.65 g/L. While the highest rhamnolipids yield of strain SG under anaerobic conditions was occurred at the 8th day. The rhamnolipids concentration in SG anaerobic culture was 0.68 g/L.
The rhlAB gene codes the rhamnosyltransferase-1 catalyzing dTDP-l-rhamnose and β-hydroxyl fatty acids to synthesis mono-rhamnolipids [17]. As shown in Fig. 1, the relative expression value of rhlAB genes was lower (0.22-fold) under anaerobic conditions than under aerobic conditions. Previous studies on the transcriptome data of P. aeruginosa also showed that the expression of rhlAB genes in P. aeruginosa was down-regulated under anaerobic conditions compared with that under aerobic conditions [29, 30]. This may explain why strain SG produced less rhamnolipids under anaerobic conditions than aerobic conditions. In our previous study, increasing the copy numbers of rhlAB genes improved rhamnolipids yield under anaerobic conditions [31], indicating that up-regulating the anaerobic expression of rhlAB genes could enhance the anaerobic production of rhamnolipids.
Fig. 1.
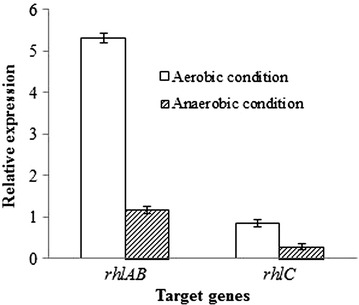
Expression of rhlAB genes and rhlC gene in P. aeruginosa SG under aerobic conditions or anaerobic conditions. The expression levels were determined by quantitative real-time PCR. 16S rRNA gene was used as endogenous control
Fourier Transform infrared spectroscopy analysis of rhamnolipids
The FT-IR spectra of two rhamnolipids products were shown in Fig. 2a, b, respectively. As shown in Fig. 2, the FT-IR spectra of two rhamnolipids products were very similar. Furthermore, both the two FT-IR spectra are similar to the reported spectra of rhamnolipids [26, 32, 33]. The characteristic absorption bands in two FT-IR spectra were explained as follows. The absorption bands around 2928, 2857, and 1457 cm−1 were, respectively, caused by the asymmetric stretching vibration of CH2, symmetric stretching vibrations of CH2, and scissoring vibrations of CH2 [33]. The absorption bands around 1731 cm−1 resulted from the ester groups. The fingerprint absorption area between 1458 and 1044 cm−1 are typical vibrations for carbohydrates. Results showed that two rhamnolipids products possessed the same structural group.
Fig. 2.
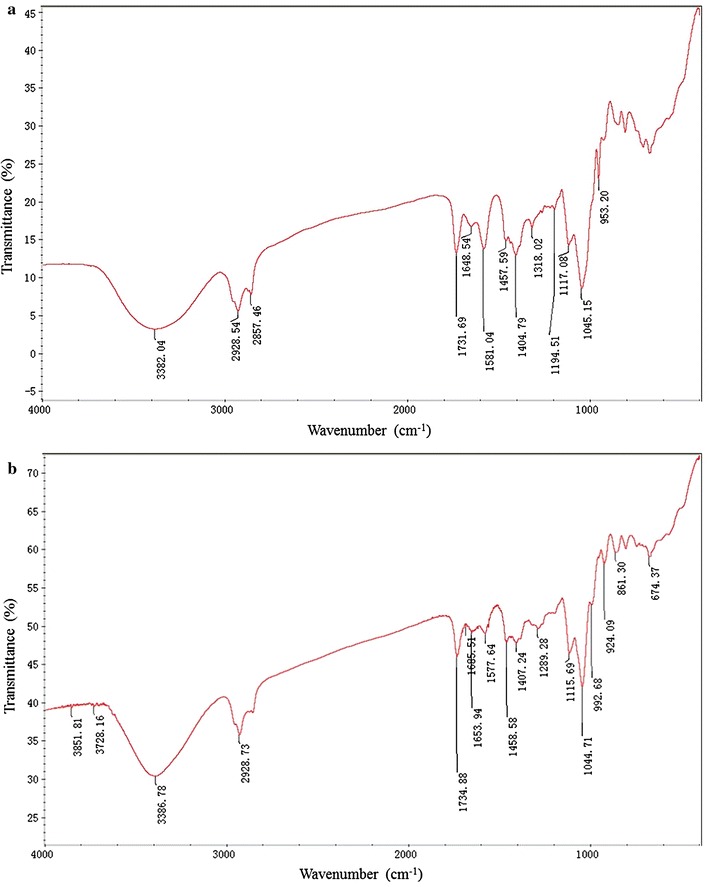
Fourier Transform infrared spectroscopy analysis of rhamnolipids products of strain SG: a rhamnolipids produced at aerobic conditions, b rhamnolipids produced at anaerobic conditions
Rhamnolipids compositions and rhlC gene expression analysis
The liquid chromatogram results of two rhamnolipids produced under aerobic conditions and anaerobic conditions were shown in Fig. 3a, b, respectively. Because the analyzed samples were the crude extracts of rhamnolipids products, there were some impurity peaks in the chromatogram. The data analysis of HPLC–MS was according to Déziel’s study [18]. As shown in Fig. 3a, there were nine rhamnolipids homologues in the rhamnolipids compound produced under aerobic conditions. But the results of Fig. 3b showed that there were seven rhamnolipids homologues in the rhamnolipids compound produced under anaerobic conditions.
Fig. 3.
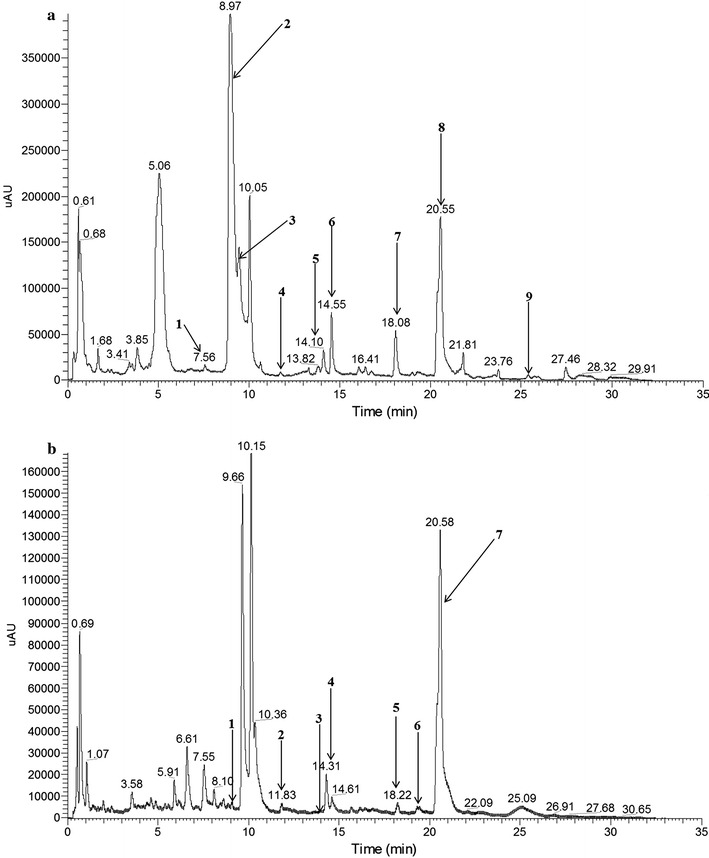
Liquid chromatogram results of rhamnolipids products of strain SG: a rhamnolipids produced at aerobic conditions, b rhamnolipids produced at anaerobic conditions
Both the two rhamnolipids products contained mono-rhamnolipids and di-rhamnolipids. As shown in Table 1, aerobically produced rhamnolipids contained three mono-rhamnolipids homologues and six di-rhamnolipids homologues. The main homologues in aerobically produced rhamnolipids are Rha–Rha-C10-C12:1, Rha–Rha-C8-C10, and Rha-C8-C10. As shown in Table 2, anaerobically produced rhamnolipids contained four mono-rhamnolipids homologues and three di-rhamnolipids homologues. The main homologues in anaerobically produced rhamnolipids are Rha-C10-C12 and Rha-C10-C10. Results confirmed that the structural compositions of two kinds of rhamnolipids products were different. According to previous studies, the concentration of rhamnolipid congeners is directly proportional to the relative abundance (%) obtained from HPLC–MS [18]. The ratio of mono-rhamnolipids to di-rhamnolipids (94.7%/4.3%) in anaerobically produced rhamnolipids was higher than that (54.8%/45.2%) in aerobically produced rhamnolipids.
Table 1.
Structural composition of rhamnolipids produced by strain SG at aerobic conditions
| Chromatographic peak number | Retention time (min) | Mass spectrum signal (m/z) | Rhamnolipids homologues | Relative abundance (%) |
|---|---|---|---|---|
| 1 | 7.56 | 447 | Rha-C8-C8 | 1.03 |
| 2 | 8.97 | 475 | Rha-C8-C10 | 52.45 |
| 3 | 9.45 | 621 | Rha–Rha-C8-C10 | 17.28 |
| 4 | 11.75 | 647 | Rha–Rha-C8-C12:1 | 0.63 |
| 5 | 14.10 | 503 | Rha-C10-C10 | 1.28 |
| 6 | 14.55 | 649 | Rha–Rha-C10-C10 | 3.32 |
| 7 | 18.08 | 677 | Rha–Rha-C10-C12 | 3.49 |
| 8 | 20.55 | 675 | Rha–Rha-C10-C12:1 | 20.03 |
| 9 | 25.38 | 705 | Rha–Rha-C12-C12 | 0.49 |
Table 2.
Structural composition of rhamnolipids produced by strain SG at anaerobic conditions
| Chromatographic peak number | Retention time (min) | Mass spectrum signal (m/z) | Rhamnolipids homologues | Relative abundance (%) |
|---|---|---|---|---|
| 1 | 9.02 | 475 | Rha-C8-C10 | 1.63 |
| 2 | 11.83 | 647 | Rha–Rha-C8-C12:1 | 1.53 |
| 3 | 14.17 | 649 | Rha–Rha-C10-C10 | 0.69 |
| 4 | 14.31 | 503 | Rha-C10-C10 | 7.13 |
| 5 | 18.22 | 677 | Rha–Rha-C10-C12 | 3.09 |
| 6 | 19.31 | 529 | Rha-C10-C12:1 | 2.66 |
| 7 | 20.58 | 531 | Rha-C10-C12 | 83.27 |
As shown in Fig. 1, the relative expression values of rhlC gene were lower (0.33-fold) under anaerobic conditions than under aerobic conditions. The rhlC gene, coding rhamnosyltransferase-2 that catalyzed mono-rhamnolipids to di-rhamnolipids, was down-regulated under anaerobic conditions, which may cause higher proportion of mono-rhamnolipids (94.7%) in anaerobically produced rhamnolipids than that (54.8%) in aerobically produced rhamnolipids.
Critical micelle concentrations and surface activity
The aerobically produced rhamnolipids and the anaerobically produced rhamnolipids decreased the air–water surface tension from 72.3 to 27.9 mN/m and 33.1 mN/m, respectively. CMC is an important parameter to assess the interfacial activity of surfactants. Surface tension of surfactants reaches the lowest value at its CMC. As shown in Fig. 4, with the increase of rhamnolipids concentrations, the surface tension values rapidly decreased. Then the surface tension remained the minimum value with the continuous increase of rhamnolipids concentration. As shown in Fig. 4a, the surface tension of solutions of aerobically produced rhamnolipids reached the minimum value (28.3 mN/m) at the concentration of 60 mg/L, which revealed the CMC of aerobically produced rhamnolipids was 60 mg/L. In Fig. 4b, the surface tension of solutions of anaerobically produced rhamnolipids reached the minimum value (33.2 mN/m) at the concentration of 80 mg/L. The CMC of anaerobically produced rhamnolipids was 80 mg/L. While the chemical surfactants sodium dodecyl sulfate (SDS) has a CMC value of 2100 mg/L [27]. The CMC of rhamnolipids produced by other bacteria ranged from 10 to 230 mg/L [26, 27, 34]. The CMC represent the surface activity of surfactants. Both the CMC values of the aerobically produced rhamnolipids and the anaerobically produced rhamnolipids were lower than the CMC values of general chemical surfactants for 2 orders of magnitude, which showed better surface activity of biosurfactants. Surfactants with lower CMC values would reach the lowest surface tension with lower surfactant concentration, which also means the surfactant can change the surface property such as emulsification and foaming even at a relatively lower concentration [27].
Fig. 4.
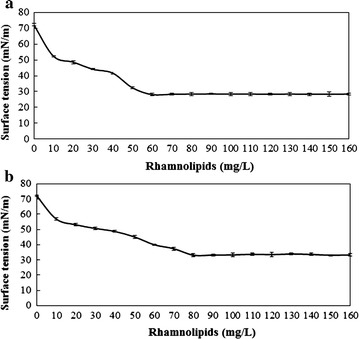
Relationship graphs between the surface tension and concentration of rhamnolipids products: a rhamnolipids produced at aerobic conditions, b rhamnolipids produced at anaerobic conditions
Emulsifying activity
Emulsifying activity of two rhamnolipids products were investigated using various hydrophobic organic compounds (including kerosene, liquid paraffin, petroleum ether, isooctane, cyclohexane, n-hexane, benzene, olive oil and crude oil). As shown in Fig. 5, both the two rhamnolipids products showed good emulsifying activity to all the tested hydrophobic organic compounds (all the EI24 > 56%). Anaerobically produced rhamnolipids has better emulsifying activity (EI24 = 80.3%) to crude oil than that (EI24 = 62.3%) of aerobically produced rhamnolipids. Using different hydrophobic organic compounds, rhamnolipids EI24 values were ranged from 53 to 90% [26, 27, 32]. Crude oil contains more complex components than other hydrophobic organic compounds, such as some components with molecular structure similar to rhamnolipids, which makes the rhamnolipids products show relative evident emulsifying activity for crude oil [35, 36]. The different structural composition makes the discrepancy in emulsifying activity for crude oil between aerobically and anaerobically produced rhamnolipids.
Fig. 5.
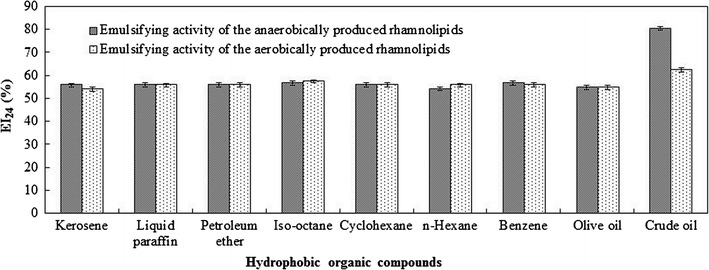
Emulsifying activity of the two types of rhamnolipids produced at aerobic or anaerobic conditions
The good emulsifying activity of the rhamnolipids products makes them suitable for bioremediation of petroleum-contaminated sites and application in microbial enhanced oil recovery (MEOR). In petroleum-contaminated environments, the presence of rhamnolipids can increase the bioavailability of the petroleum hydrocarbons through emulsification [9, 37, 38]. Furthermore, rhamnolipids can be used as oil-displacing agent to mobilize the trapped oil in reservoirs based on its good oil emulsification activity [7, 39–41].
Stability of two rhamnolipids products
Two rhamnolipids products were treated at different temperatures, pH and salinity. The surface tension and EI24 index of treated rhamnolipids solutions were measured at 35 °C to evaluate the stability of two rhamnolipids products. Two rhamnolipids products still reduced the surface tension of water to lower than 31 and 35.0 mN/m, respectively, after treated at temperatures (4–121 °C), pH values (2–10) and 0–150 g/L of NaCl. Two rhamnolipids products both had EI24 values (against crude oil) higher than 60% after treated at temperatures (4–121 °C), pH values (2–10) and 0–90 g/L of NaCl. There was no difference in the EI24 values among the different conditions. Results revealed that two rhamnolipids products were stable at different temperatures, pH and salinity. Although the structural compositions of two rhamnolipids products were different, both the two rhamnolipids products were thermo-stable and salt-tolerant. Both two rhamnolipids products are promising for biotechnology use in the complex and extreme environments, such as oil reservoirs, river sediment and petroleum contaminated soil.
Application perspectives
Rhamnolipids consists of a hydrophilic moiety (one or two rhamnoses) and a hydrophobic moiety (saturated or unsaturated β-hydroxy fatty acids with different carbon chain lengths) [17]. Because of containing only one rhamnose, mono-rhamnolipids exhibits weaker hydrophilic ability and stronger lipophilic capability than di-rhamnolipids, i.e. mono-rhamnolipids has better emulsifying activity than di-rhamnolipids. Previous study reported that di-rhamnolipids was more effective than mono-rhamnolipids for desorption of triclosan from sediments [42], which confirmed that di-rhamnolipids had stronger surface activity than mono-rhamnolipids.
In this study, the ratio of mono-rhamnolipids to di-rhamnolipids (94.7%/4.3%) in anaerobically produced rhamnolipids was higher than that (54.8%/45.2%) in aerobically produced rhamnolipids. Therefore, the anaerobically produced rhamnolipids exhibited better emulsifying activity, but the aerobically produced rhamnolipids showed better surface activity, such as lower CMC value and lower surface tension. The unique properties of the anaerobically produced rhamnolipids validated strain P. aeruginosa SG as a promising biosurfactants producer for MEOR applications and environmental remediation of hydrocarbon pollutants. However, strain P. aeruginosa SG produced less rhamnolipids under anaerobic conditions than aerobic conditions. Therefore, future research would concentrate on how to enhance anaerobic production of rhamnolipids, such as using strain mutation, biosynthesis pathway regulation and gene manipulation. In future, we can regulate microorganisms to synthesize highly active rhamnolipids according to different biotechnology application requirements.
Rhamnolipids can improve the flow ability of trapped oil in the subsurface oil reservoirs and assist in the detachment of oil films from rocks based on its good oil emulsification activity [39, 40]. Furthermore, rhamnolipids can increase the bioavailability of the petroleum hydrocarbons through its emulsifying activity [9, 37, 38]. Anaerobically produced rhamnolipids of strain SG exhibited better emulsifying activity. Therefore, the anaerobically produced rhamnolipids would be more advantageous for applications in enhanced oil recovery and bioremediation of petroleum contaminated sites.
Conclusions
Oxygen conditions affected the rhl-genes expression in P. aeruginosa SG, thus altering the rhamnolipids yield, structural composition and physicochemical properties. Rhamnolipids produced at aerobic or anaerobic conditions had distinct application potential in diverse biotechnologies. The rhlAB and rhlC genes were down-regulated under anaerobic conditions. Strain SG produced less rhamnolipids under anaerobic conditions than under aerobic conditions. Rhamnolipids produced at anaerobic conditions contained more mono-rhamnolipids. Aerobically produced rhamnolipids possesed better surface activity, so it is advantageous in the wetting and desorption applications. Anaerobically produced rhamnolipids exhibited better emulsifying activity, so it is promising for applied in MEOR and petroleum pollution bioremediation.
Authors’ contributions
FZ carried out the experiments, analyzed the data and drafted the manuscript. YZ conceived and supervised the study and reviewed the final manuscript. SQH assisted in bacterial cultivation experiments. RJS and FM participated in the design of the study and structural analysis of rhamnolipids. All authors read and approved the final manuscript.
Acknowledgements
This work was financially supported by the National Natural Science Foundation of China (31700117), the China Postdoctoral Science Foundation (2017M621292) and the Open Fund of Key Laboratory of Pollution Ecology and Environmental Engineering, Chinese Academy of Sciences.
Competing interests
The authors declare that they have no competing interests.
Availability of data and materials
The datasets supporting the conclusions of this article are included within the article.
Consent for publication
All the co-authors approved the publication of this work in Microbial Cell Factories.
Ethics approval and consent to participate
Not applicable.
Funding
This work was financially supported by the National Natural Science Foundation of China (31700117) and the China Postdoctoral Science Foundation (2017M621292).
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Feng Zhao, Email: zhaofeng@iae.ac.cn.
Rongjiu Shi, Email: shirongjiu@iae.ac.cn.
Fang Ma, Email: mafangzfhit@126.com.
Siqin Han, Email: hansq@iae.ac.cn.
Ying Zhang, Email: yzhang@iae.ac.cn.
References
- 1.Müller MM, Kügler JH, Henkel M, Gerlitzki M, Hörmann B, Pöhnlein M, Syldatk C, Hausmann R. Rhamnolipids-next generation surfactants? J Biotechnol. 2012;162:366–380. doi: 10.1016/j.jbiotec.2012.05.022. [DOI] [PubMed] [Google Scholar]
- 2.De S, Malik S, Ghosh A, Saha R, Saha B. A review on natural surfactants. RSC Adv. 2015;5(81):65757–65767. doi: 10.1039/C5RA11101C. [DOI] [Google Scholar]
- 3.Chong H, Li Q. Microbial production of rhamnolipids: opportunities, challenges and strategies. Microb Cell Fact. 2017;16(1):137. doi: 10.1186/s12934-017-0753-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Santos DKF, Rufino RD, Luna JM, Santos VA, Sarubbo LA. Biosurfactants: multifunctional biomolecules of the 21st century. Int J Mol Sci. 2016;17(3):401. doi: 10.3390/ijms17030401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Amani H, Müller MM, Syldatk C, Hausmann R. Production of microbial rhamnolipids by Pseudomonas aeruginosa MM1011 for ex situ enhanced oil recovery. Appl Biochem Biotechnol. 2013;170:1080–1093. doi: 10.1007/s12010-013-0249-4. [DOI] [PubMed] [Google Scholar]
- 6.Brown LR. Microbial enhanced oil recovery (MEOR) Curr Opin Microbiol. 2010;13:316–320. doi: 10.1016/j.mib.2010.01.011. [DOI] [PubMed] [Google Scholar]
- 7.Gudiña EJ, Rodrigues AI, Alves E, Domingues MR, Teixeira JA, Rodrigues LR. Bioconversion of agro-industrial by-products in rhamnolipids toward applications in enhanced oil recovery and bioremediation. Bioresour Technol. 2015;177:87–93. doi: 10.1016/j.biortech.2014.11.069. [DOI] [PubMed] [Google Scholar]
- 8.Cameotra SS, Singh P. Synthesis of rhamnolipid biosurfactant and mode of hexadecane uptake by Pseudomonas species. Microb Cell Fact. 2009;8(1):16. doi: 10.1186/1475-2859-8-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Whang LM, Liu PWG, Ma CC, Cheng SS. Application of biosurfactant, rhamnolipids, and surfactin, for enhanced biodegradation of diesel-contaminated water and soil. J Hazard Mater. 2008;151:155–163. doi: 10.1016/j.jhazmat.2007.05.063. [DOI] [PubMed] [Google Scholar]
- 10.Zou H, Du W, Ji M, Zhu R. Enhanced electrokinetic remediation of pyrene-contaminated soil through pH control and rhamnolipids addition. Environ Eng Sci. 2016;33(7):507–513. doi: 10.1089/ees.2016.0019. [DOI] [Google Scholar]
- 11.Abdel-Mawgoud AM, Lepine F, Deziel E. Rhamnolipids: diversity of structures, microbial origins and roles. Appl Microbiol Biotechnol. 2010;86:1323–1336. doi: 10.1007/s00253-010-2498-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shekhar S, Sundaramanickam A, Balasubramanian T. Biosurfactant producing microbes and their potential applications: a review. Crit Rev Environ Sci Technol. 2015;45(14):1522–1554. doi: 10.1080/10643389.2014.955631. [DOI] [Google Scholar]
- 13.Arai H. Regulation and function of versatile aerobic and anoxic respiratory metabolism in Pseudomonas aeruginosa. Front Microbiol. 2011;2:1–13. doi: 10.3389/fmicb.2011.00103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schobert M, Jahn D. Anaerobic physiology of Pseudomonas aeruginosa in the cystic fibrosis lung. Int J Med Microbiol. 2010;300:549–556. doi: 10.1016/j.ijmm.2010.08.007. [DOI] [PubMed] [Google Scholar]
- 15.Chayabutra C, Wu J, Ju LK. Rhamnolipids production by Pseudomonas aeruginosa under denitrification: effects of limiting nutrients and carbon substrates. Biotechnol Bioeng. 2001;72:25–33. doi: 10.1002/1097-0290(20010105)72:1<25::AID-BIT4>3.0.CO;2-J. [DOI] [PubMed] [Google Scholar]
- 16.Zhao F, Zhang J, Shi R, Han S, Ma F, Zhang Y. Production of biosurfactant by a Pseudomonas aeruginosa isolate and its applicability to in situ microbial enhanced oil recovery under anoxic conditions. RSC Adv. 2015;5(45):36044–36050. doi: 10.1039/C5RA03559G. [DOI] [Google Scholar]
- 17.Soberón-Chávez G, Lepine F, Déziel E. Production of rhamnolipids by Pseudomonas aeruginosa. Appl Microbiol Biotechnol. 2005;68:718–725. doi: 10.1007/s00253-005-0150-3. [DOI] [PubMed] [Google Scholar]
- 18.Déziel E, Lépine F, Dennie D, Boismenu D, Mamer OA, Villemur R. Liquid chromatography/mass spectrometry analysis of mixtures of rhamnolipids produced by Pseudomonas aeruginosa strain 57RP grown on mannitol or naphthalene. Biochim Biophys Acta Mol Cell Biol Lipids. 1999;1440(2):244–252. doi: 10.1016/S1388-1981(99)00129-8. [DOI] [PubMed] [Google Scholar]
- 19.Nicolò MS, Cambria MG, Impallomeni G, Rizzo MG, Pellicorio C, Ballistreri A, Guglielmino SP. Carbon source effects on the mono/dirhamnolipid ratio produced by Pseudomonas aeruginosa L05, a new human respiratory isolate. New Biotechnol. 2017;39:36–41. doi: 10.1016/j.nbt.2017.05.013. [DOI] [PubMed] [Google Scholar]
- 20.Han L, Liu P, Peng Y, Lin J, Wang Q, Ma Y. Engineering the biosynthesis of novel rhamnolipids in Escherichia coli for enhanced oil recovery. J Appl Microbiol. 2014;117(1):139–150. doi: 10.1111/jam.12515. [DOI] [PubMed] [Google Scholar]
- 21.Müller MM, Hausmann R. Regulatory and metabolic network of rhamnolipid biosynthesis: traditional and advanced engineering towards biotechnological production. Appl Microbiol Biotechnol. 2011;91(2):251–264. doi: 10.1007/s00253-011-3368-2. [DOI] [PubMed] [Google Scholar]
- 22.Zhao F, Zhou J, Han S, Ma F, Zhang Y, Zhang J. Medium factors on anaerobic production of rhamnolipids by Pseudomonas aeruginosa SG and a simplifying medium for in situ microbial enhanced oil recovery applications. World J Microbiol Biotechnol. 2016;32(4):54. doi: 10.1007/s11274-016-2020-9. [DOI] [PubMed] [Google Scholar]
- 23.Guyard-Nicodème M, Bazire A, Hémery G, Meylheuc T, Mollé D, Orange N, Chevalier S. Outer membrane modifications of Pseudomonas fluorescens MF37 in response to hyperosmolarity. J Proteome Res. 2008;7(3):1218–1225. doi: 10.1021/pr070539x. [DOI] [PubMed] [Google Scholar]
- 24.Bazire A, Dheilly A, Diab F, Morin D, Jebbar M, Haras D, Dufour A. Osmotic stress and phosphate limitation alter production of cell-to-cell signal molecules and rhamnolipid biosurfactant by Pseudomonas aeruginosa. FEMS Microbiol Lett. 2005;253(1):125–131. doi: 10.1016/j.femsle.2005.09.029. [DOI] [PubMed] [Google Scholar]
- 25.Neilson JW, Zhang L, Veres-Schalnat TA, Chandler KB, Neilson CH, Crispin JD, Pemberton JE, Maier RM. Cadmium effects on transcriptional expression of rhlB/rhlC genes and congener distribution of monorhamnolipid and dirhamnolipid in Pseudomonas aeruginosa IGB83. Appl Microbiol Biotechnol. 2010;88(4):953–963. doi: 10.1007/s00253-010-2808-8. [DOI] [PubMed] [Google Scholar]
- 26.Xia WJ, Luo ZB, Dong HP, Yu L, Cui QF, Bi YQ. Synthesis, characterization, and oil recovery application of biosurfactant produced by indigenous Pseudomonas aeruginosa WJ-1 using waste vegetable oils. Appl Biochem Biotechnol. 2012;166:1148–1166. doi: 10.1007/s12010-011-9501-y. [DOI] [PubMed] [Google Scholar]
- 27.Yin H, Qiang J, Jia Y, Ye JS, Peng H, Qin HM, Zhang N, He BY. Characteristics of biosurfactant produced by Pseudomonas aeruginosa S6 isolated from oil-containing wastewater. Process Biochem. 2009;44:302–308. doi: 10.1016/j.procbio.2008.11.003. [DOI] [Google Scholar]
- 28.Zhao F, Liang X, Ban Y, Han S, Zhang J, Zhang Y, Ma F. Comparison of methods to quantify rhamnolipids and optimization of oil spreading method. Tenside Surfact Det. 2016;53(3):243–248. doi: 10.3139/113.110429. [DOI] [Google Scholar]
- 29.Alvarez-Ortega C, Harwood CS. Responses of Pseudomonas aeruginosa to low oxygen indicate that growth in the cystic fibrosis lung is by aerobic respiration. Mol Microbiol. 2007;65(1):153–165. doi: 10.1111/j.1365-2958.2007.05772.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Filiatrault MJ, Wagner VE, Bushnell D, Haidaris CG, Iglewski BH, Passador L. Effect of anaerobiosis and nitrate on gene expression in Pseudomonas aeruginosa. Infect Immun. 2005;73(6):3764–3772. doi: 10.1128/IAI.73.6.3764-3772.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhao F, Cui Q, Han S, Dong H, Zhang J, Ma F, Zhang Y. Enhanced rhamnolipids production of Pseudomonas aeruginosa SG by increasing copy number of rhlAB genes with modified promoter. RSC Adv. 2015;5(86):70546–70552. doi: 10.1039/C5RA13415C. [DOI] [Google Scholar]
- 32.Albino JD, Namb IM. Partial characterization of biosurfactant produced under anaerobic conditions by Pseudomonas sp. ANBIOSURF-1. Adv Mater Res. 2010;93:623–626. doi: 10.4028/www.scientific.net/AMR.93-94.623. [DOI] [Google Scholar]
- 33.Kiefer J, Radzuan MN, Winterburn J. Infrared spectroscopy for studying structure and aging effects in rhamnolipid biosurfactants. Appl Sci. 2017;7(5):533. doi: 10.3390/app7050533. [DOI] [Google Scholar]
- 34.Nitschke M, Cost SGVAO, Contiero J. Rhamnolipid surfactants: an update on the general aspects of these remarkable biomolecules. Biotechnol Prog. 2005;21(6):1593–1600. doi: 10.1021/bp050239p. [DOI] [PubMed] [Google Scholar]
- 35.Long X, Zhang G, Shen C, Sun G, Wang R, Yin L, Meng Q. Application of rhamnolipid as a novel biodemulsifier for destabilizing waste crude oil. Bioresour Technol. 2013;131:1–5. doi: 10.1016/j.biortech.2012.12.128. [DOI] [PubMed] [Google Scholar]
- 36.Pekdemir T, Copur M, Urum K. Emulsification of crude oil–water systems using biosurfactants. Process Saf Environ. 2005;83(1):38–46. doi: 10.1205/psep.03176. [DOI] [Google Scholar]
- 37.Rahman KSM, Rahman TJ, Kourkoutas Y, Petsas I, Marchant R, Banat IM. Enhanced bioremediation of n-alkane in petroleum sludge using bacterial consortium amended with rhamnolipids and micronutrients. Bioresour Technol. 2003;90(2):159–168. doi: 10.1016/S0960-8524(03)00114-7. [DOI] [PubMed] [Google Scholar]
- 38.Sponza DT, Gök O. Effect of rhamnolipids on the aerobic removal of polyaromatic hydrocarbons (PAHs) and COD components from petrochemical wastewater. Bioresour Technol. 2010;101(3):914–924. doi: 10.1016/j.biortech.2009.09.022. [DOI] [PubMed] [Google Scholar]
- 39.Wang Q, Fang X, Bai B, Liang X, Shuler PJ, Goddard WA, III, Tang Y. Engineering bacteria for production of rhamnolipid as an agent for enhanced oil recovery. Biotechnol Bioeng. 2007;98:842–853. doi: 10.1002/bit.21462. [DOI] [PubMed] [Google Scholar]
- 40.Varjani SJ, Upasani VN. Core flood study for enhanced oil recovery through ex situ bioaugmentation with thermo- and halo-tolerant rhamnolipids produced by Pseudomonas aeruginosa NCIM 5514. Bioresour Technol. 2016;220:175–182. doi: 10.1016/j.biortech.2016.08.060. [DOI] [PubMed] [Google Scholar]
- 41.Hosseininoosheri P, Lashgari HR, Sepehrnoori K. A novel method to model and characterize in situ bio-surfactant production in microbial enhanced oil recovery. Fuel. 2016;183:501–511. doi: 10.1016/j.fuel.2016.06.035. [DOI] [Google Scholar]
- 42.Zhang X, Guo Q, Hu Y, Lin H. Effects of monorhamnolipid and dirhamnolipid on sorption and desorption of triclosan in sediment–water system. Chemosphere. 2013;90(2):581–587. doi: 10.1016/j.chemosphere.2012.08.036. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets supporting the conclusions of this article are included within the article.


