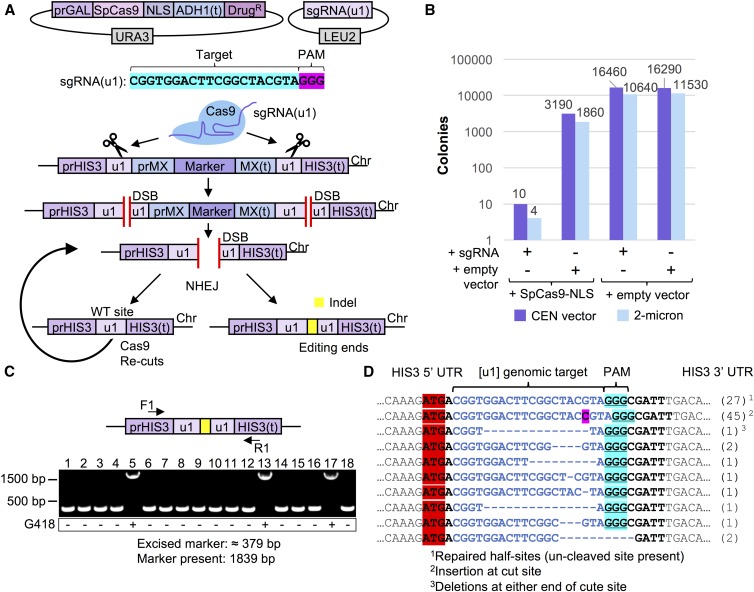
Figure 1.
A safe, programmable system to test CRISPR-based gene editing in haploid yeast. (A) Our design for a yeast system for analysis of CRISPR editing includes (i) an inducible S. pyogenes Cas9 expressed from a URA3-based plasmid, (ii) a sgRNA expression cassette on a high-copy LEU2-based plasmid, and (iii) a programmable gene “target” (consisting of a drug resistance marker cassette) at a safe-harbor locus (HIS3) flanked by two “unique” DNA sequences (u1) that do not exist within the S. cerevisiae genome (Finnigan and Thorner 2016). Induction of Cas9 allows targeting and double-stranded break formation at the identical (u1) sequences. In the absence of exogenous DNA (e.g., amplified PCR product) to be used for HDR, NHEJ of the exposed chromosomal ends causes full excision of the selectable marker. However, given the unique arrangement of the identical (u1) sites, NHEJ in the absence of any insertion/deletion mutation at the Cas9 cut site (left) recreates another WT (u1) site and subsequent re-editing of the same target sequence until Cas9 expression is shutoff or a mutation is positioned within the (u1) site (right). (B) Cas9-dependent editing results in cell inviability. GFY-2353 yeast already harboring Cas9-NLS on a vector (pGF-IVL1116) or an empty vector control (pRS316) were induced in medium containing galactose, transformed with the sgRNA(u1)-expression cassette on either a CEN-based (pGF-V1215) or 2μ-based (pGF-V1220) plasmid, and plated onto SD-URA-LEU media. (C) GFY-2588 yeast containing pGF-IVL1342 were transformed with sgRNA(u1) plasmid (pGF-V1216) and selected on SD-URA-LEU medium. The isolated chromosomal DNA of individual clonal (surviving) isolates was assayed by PCR using DNA oligonucleotides (F1/R1, Table S1 in File S1) to the flanking HIS3 UTR. The expected product sizes of the amplified PCR fragments are ∼379 bp (depending on the type of insertion/deletion(s) at the cut site, if any), or 1839 bp in the absence of any editing. Colonies were tested for resistance on medium containing G418 (below). (D) Clonal isolates from Cas9 editing (a dozen independent experiments) using the high copy sgRNA(u1) plasmid from (B) and that had also excised the selection cassette were analyzed by DNA sequencing at the HIS3 locus. The number of each genotype obtained is listed (right).