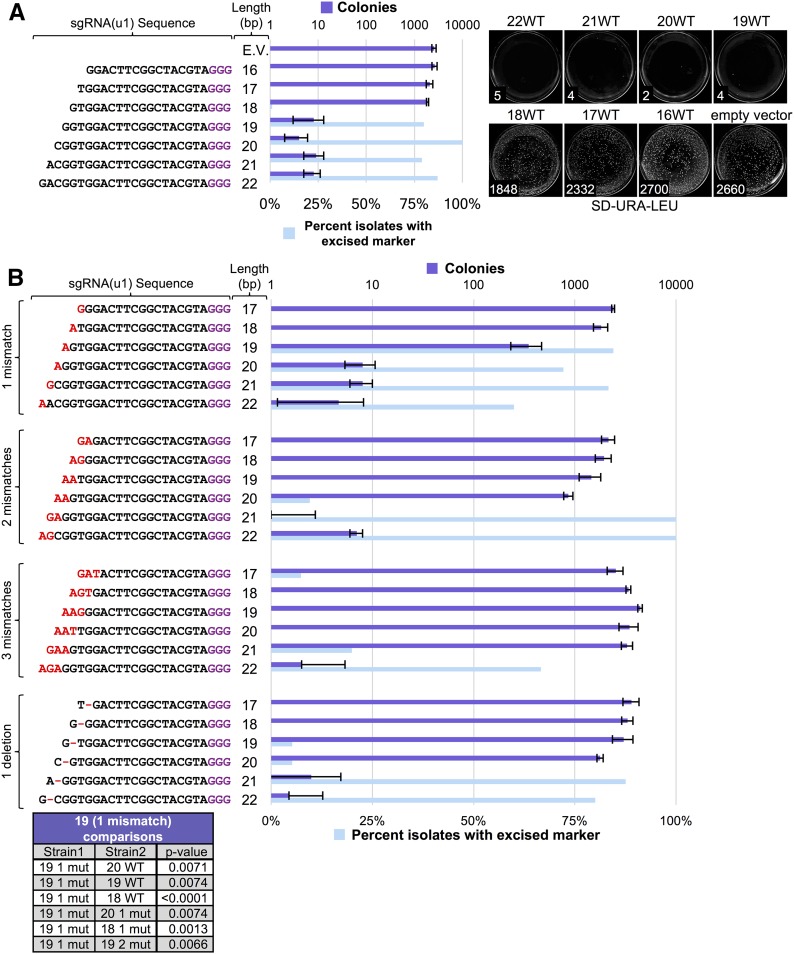
Figure 2.
Effect of sgRNA length and 5′ target mismatch on Cas9 editing efficiency. (A) GFY-2353 yeast containing the Cas9-NLS vector (pGF-IVL1116) were transformed with sgRNA(u1) cassettes (plasmids pGF-V1216–pGF-V1222) with guide sequences of varying length along with an empty pRS425 vector control. The number of colonies was quantified (left) for three independent trials. Error, SD. Representative plates are shown (right). A random sampling of colonies was chosen across all three trials following editing on SD-URA-LEU plates and tested for growth on rich medium containing hygromycin. The percentage of isolates displaying sensitivity to the drug were quantified. For conditions (e.g., sgRNA(u1) 20 bp length) where a small number of colonies were viable, all surviving isolates (typically 5–20 total) were tested on hygromycin; for other combinations, between 150 and 200 colonies were sampled. (B) Cas9 editing was repeated as in (A) using sgRNA(u1) cassettes containing varying mismatches at the 5′ end of the guide sequence. A single mismatch at the 5′ end (pGF-V1223 – pGF-V1228), two mismatches (pGF-V1229 – pGF-V1234), three mismatches (pGF-V1235 – pGF-V1240), or a deletion of one base at the penultimate −2 position from the 5′ end (pGF-V1241–pGF-V1246) were assayed for both total surviving colonies and the percentage of isolates with an excised marker cassette at the target locus (top). Select comparisons with the sgRNA(u1) 19 bp guide with one mismatch data were performed using an unpaired t-test (bottom).