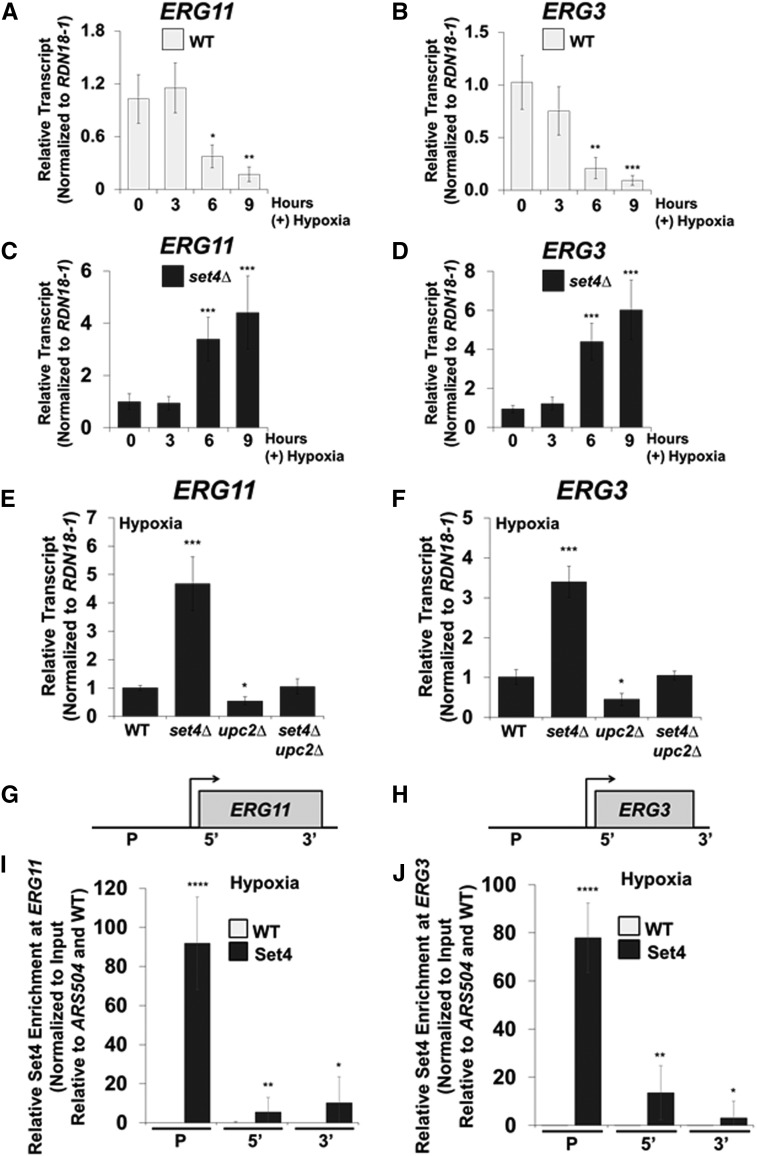
Figure 5.
Set4 represses ergosterol genes and directly targets the promoters of ERG11 and ERG3 under hypoxia. (A and B) The mRNA transcript levels of ERG11 and ERG3 were determined in WT cells grown under aerobic or hypoxic conditions over time. qRT-PCR expression analysis in the WT strain was set relative to the aerobic or hypoxic WT using the 18S ribosome rRNA (RDN18-1) as the internal control to normalize transcript levels. (C and D) qRT-PCR analysis of ERG11 and ERG3 expression in set4Δ cells grown under aerobic or hypoxic conditions for 3, 6, or 9 hr. ERG11 and ERG3 expression in the set4∆ strain were normalized to RDN18-1 and set to the WT strain at the same relative time point. Data were analyzed from three biological replicates that had three technical replicates each. Error bars represent SD. * P < 0.05, ** P < 0.01, *** P < 0.005. (E and F) Gene expression analysis (qRT-PCR) of ERG11 and ERG3 under hypoxic conditions in WT, set4Δ, upc2Δ, and set4Δupc2Δ. The mRNA transcript levels were normalized to RDN18-1 and set relative to WT grown under hypoxia. * P < 0.05, *** P < 0.005. (G and H) Schematics of ERG11 and ERG3 loci with the specified positions of ChIP probes. (I and J) ChIP analysis of Set4 enriched at ERG11 and ERG3 was performed under hypoxic conditions using antibodies specific to a 3×FLAG tag for the detection of Set4. ChIP analyses were normalized to DNA input samples and set relative to the ARS504 and untagged WT. Error bars represent SD for three biological replicates with three technical replicates each. * P < 0.05, ** P < 0.01, **** P < 0.0001. Gene expression and ChIP analyses were performed using the FY2609 strain.